Analyzing High-Resolution Microscopy Images
By Roy Wollman Ph.D., University of California, Davis
Automated microscopy enables the acquisition of high-resolution images at rates of one image per second and total numbers in the millions. These high rates pose a new challenge in cell biology: how to analyze the vast amounts of resulting data in a reasonable time frame?
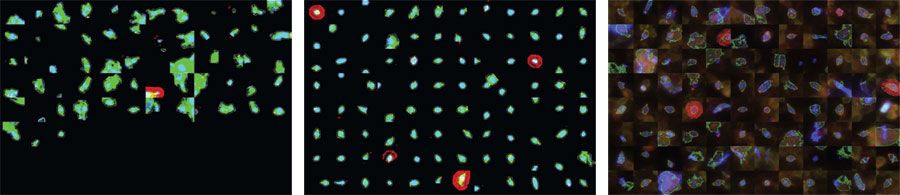
My colleagues and I confronted this challenge when we analyzed cells of the Drosophila fruit fly during mitosis. Our goal was to understand the genes involved in normal and abnormal cell division (Figure 1). Since cancer is, in many respects, uncontrolled cell division, chemotherapies often target cells during mitosis. Identifying new genes involved in mitosis could open the way to new targets for chemotherapeutic drugs.
Our project, a collaboration between the Vale lab at UC San Francisco and the Scholey lab at UC Davis, involved performing an image-based, genome-wide RNAi screen of Drosophila cells. Traditional tools used by cell biologists, which rely heavily on visual inspection, do not scale well, and cannot be used for large-scale projects. High-throughput microscopy was crucial due to the high rate of acquisition and the low percent of dividing cells in the population. Because we screen only for the 1% of cells that are dividing, we need to take 100 times more images.
We used MATLAB®, Image Processing Toolbox™, and Statistics and Machine Learning Toolbox™ to develop routines that could process these images straight from the microscope, keeping up with the rate of acquisition when running four CPUs in real time.
High-Resolution Microscopy
High-resolution microscopy is the standard ”readout” for functional assays in cell biology. It provides visual information on subcellular organelles, specialized organs of the cell that are only a few microns in size. In fluorescence microscopy, the sample is stained with markers of a few (usually not more than five) key molecules. These markers reveal information on the properties of the organelles, including their shape, geometry, intensity, and relation to other structures.
In a typical cell biology experiment, a perturbation, (a manipulation to the regular state of the cell) is performed, and the properties of the cells when in a perturbed and a non-perturbed state are compared. The results help biologists understand what the perturbation affected mechanistically and therefore, learn more about how cells function. Technological developments in functional genomics enable these perturbations to be performed on a scale never before seen, generating data at unprecedented rates.
RNA Interference
RNA interference (RNAi) is a methodology that piggy-backs on the cell’s own machinery and can reduce the genetic expression of every gene in the genome by more than 90%. Labs around the world have developed libraries of RNAi probes targeting every gene in the genome of multiple model organisms. These new tools let researchers test on a genome-wide scale which genes are involved in a particular biological process of interest.
Image-based RNAi screens combine systematic perturbation with automated microscopy that performs rapid acquisition (rates of ~1 image per second) of high-resolution images. A typical RNAi library contains ~25,000 probes. The acquisition of 10 images per probe with 4 different stains results in 1,000,000 images that need to be analyzed.
Analyzing the Images
The images were entered straight from the microscope into a database, where they were constantly monitored by a custom bash daemon that initiated our MATLAB based analysis procedure. The image analysis involved five steps:
- Segmenting the images to identify all the cells in an image
- Classifying the cells as mitotic (in the middle of division) or interphase (non-dividing)
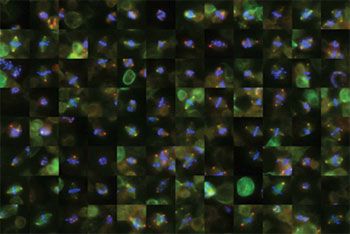
- Creating galleries of cells during division to enable rapid visual inspection
- Performing quantitative measurements on these cells
- Using bootstrap statistics to identify statistically significant hits (perturbation that caused changes to the phenotypes higher than expected by chance)
Image Processing Toolbox provides implementations of all the basic image manipulation tools (Figure 2). These tools were the building blocks of our analysis procedure.
MATLAB algorithms are easy to use and very well documented, allowing scientists who are not expert programmers to tweak parameters. By tuning and combining these existing algorithms we were able to focus on the problem at hand and not get bogged down in programming and implementation details.
We also made extensive use of Netlab, a library of artificial neural networks, one of the many open-source toolboxes available from the large MATLAB user community. With its well developed external interface and APIs, MATLAB enabled us to combine Netlab with other tools and languages to create a well integrated analysis pipeline.
Meeting the Challenges of Real-Time Screening
In the middle of our screening procedure, the images started to look different. We suspected that, probably due to a change in the medium used to feed the cells, an internal change in cell behavior had caused one of the markers that we were using to lose specificity. As a result, our classification procedure stopped working properly, and we had to adapt it rapidly while the images were streaming. This required retraining the neural network classifier that we use based on the new sets of images coming from the microscope. We used MATLAB visualization tools to quickly identify what had gone wrong and adapt our analysis procedure accordingly.
The nature of scientific discovery is that you don’t always know in advance what exactly you are looking for. Toward the end of the screening process, the cell biologist who visually inspected thousands of galleries suspected that we were missing a potentially important phenotype. We used MATLAB to develop additional assays in a matter of days. We included them in the analysis pipeline without stopping the acquisition process, and added these assays to the screen. The new assays were responsible for the identification of many of the novel results that came from this study.
One of the biggest challenges in high-throughput image-based screens is the problem of the high number of false-positive hits. MATLAB and Statistics and Machine Learning Toolbox enabled us to adapt the statistical analysis using a computationally intensive re-sampling methodology and to rigorously assign a p-value for each test using nonparametric tools. The advantage of nonparametric tools is that they estimate the “wild type” null distribution for each plate separately, which tends to reduce the number of false-positives. MATLAB matrix-handling capabilities enabled us to implement the re-sampling algorithms with only a few lines of code.
Research Results
The importance of high-content, image-based screens in cell biology is only growing, and I have no doubt that researchers will continue to rely on MATLAB for image analysis in cell biology. Many other image-based screens either use MATLAB tools directly or develop libraries that are suitable for specific research in cell biology—for example, CellProfiler.
Overall, our project was successful. We identified 204 of the genes that are required for mitosis, many of them novel or unexpected. Many were further verified as important in other organisms, including humans. Follow-up research on these genes, looking into the details of their mechanism of function and how depletion of these genes affects cell division, continues in labs around the world.
Published 2008 - 91566v00