Evaluating Alzheimer’s Disease Therapeutic Targets with Deep Learning and the MATLAB Based Spine Tool
By Justin Elstrott, PhD, Department of Biomedical Imaging, Genentech Inc.
Alzheimer's disease (AD), the most common cause of dementia, is characterized by changes in the brain resulting from the buildup of amyloid plaques. Studies have shown that these hard, insoluble accumulations of beta amyloid proteins are closely linked with the loss of dendritic spines, micron-sized protrusions from a neuron’s dendrite that receive input from other neurons. These studies have led my Genentech colleagues and other AD researchers to preclinically evaluate compounds that reduce spine loss in the presences of amyloid plaques.
To quantify spine loss, we examine microscopic images of mouse brain tissue, counting individual spines along the dendrite and calculating spine density (for example, the number of spines per 100 micrometers of dendrite). When performed manually, this process is extremely labor intensive.
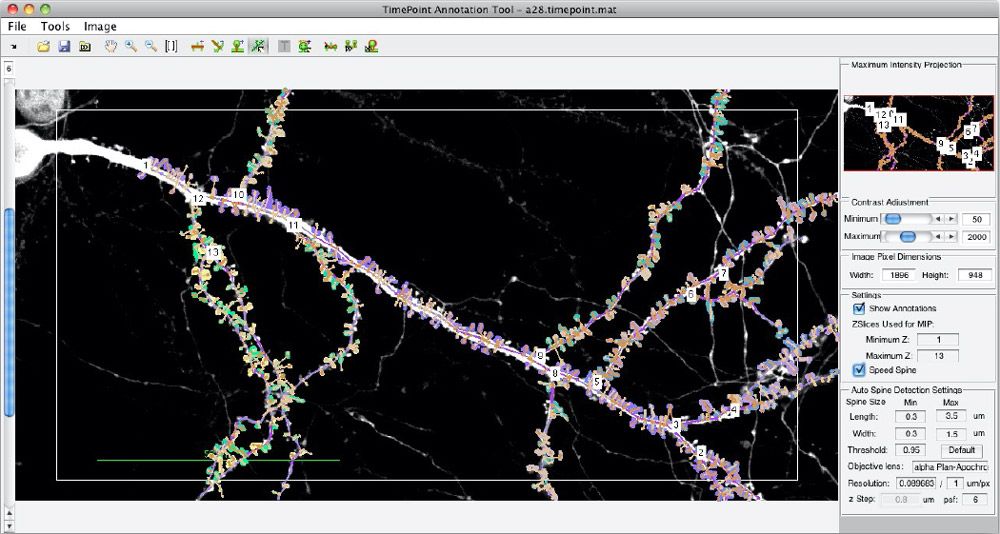
About 10 years ago, our group developed Spine Tool, a MATLAB® image processing application that helps automate spine identification and spine density calculations (Figure 1). While the original version of Spine Tool worked well for in vitro samples, it produced a nontrivial number of false positives and false negatives in ex vivo brain samples, where image quality is lower.
To improve the accuracy of Spine Tool, we worked with MathWorks consultants to incorporate deep learning into the tool. We trained a convolutional neural network (CNN) on a data set of more than 9,000 images that we had annotated with the original Spine Tool. The trained CNN has further automated the process of evaluating therapeutics for minimizing spine loss.
Spine Density Basics
To conduct our studies, we use normal mice (wild-type or WT mice) and mice that express amyloid beta (PS2APP mice), which tend to develop amyloid plaques. Using digital images of microscopic slices of mice brain tissue, we identify and count spines along individual dendrites in both the presence and the absence of visible plaques.
As Figure 2 shows, the presence of the plaque in PS2APP mice corresponds with a visible reduction in the number of nearby spines. That particular study included a third group of mice in which the complement 3 gene (C3KO) was removed; those mice demonstrated significantly less spine loss [1].

Figure 2. Mouse dendrites with spines. Left to right: a normal mouse (WT), a mouse with suppressed expression of the C3 gene (C3KO), a PS2APP mouse (with the dendrites away from and near a plaque), and a PS2APP and C3KO mouse (with the dendrites away from and near a plaque). Adapted from [1].
We aggregate the spine densities from hundreds of samples to produce a bar chart that compares the densities for different mouse genotypes (Figure 3), revealing that both the reduction in spine density near plaques and the recovery of spine density in C3KO mice are statistically significant. These study results suggest that reducing complement activity could be an effective therapeutic strategy.

Figure 3. Chart showing averaged spine density for various genotypes. Adapted from [1].
Combining Traditional Image Processing with Deep Learning
Earlier versions of Spine Tool identified dendritic spines by applying threshold detection, segmentation, and morphological image processing techniques, as well as custom operations for detecting neuroscience-specific morphologies (Figure 4).
To incorporate deep learning into Spine Tool, we evaluated several predefined network architectures, including Deeplab, SegNet, and U-Net, a network developed for biomedical image segmentation. We chose U-Net for its multiresolution performance.
To reduce training time, we decided to use the 2D version of U-Net rather than the more complex 3D version. Although the data we work with is three-dimensional, it is not isometric: the volumes are only a few slices deep, and a dendritic spine rarely spans more than one or two slices along the Z axis. We created 2D data sets by performing maximum intensity projection of the thin 3D slices. We saved additional time by performing the training on a workstation with NVIDIA® GeForce RTX 2080 Ti GPUs using Parallel Computing Toolbox™.
Initial classification results from the trained network were promising but still included false negatives, identifying fewer spines than we could see ourselves. This was due to a class imbalance problem: the spines are tiny in comparison to the dendrites and the background. We adjusted the class weights across the network and evaluated various loss functions to improve prediction accuracy. We opted to set the weights to maximize sensitivity.
The updated network detected virtually all the spines in an image, but with more false positives. We eliminated these false positive by post-processing the results. For example, we applied length and volume thresholds on the detected spines to weed out any that were too large or too small to be actual spines. To date, our deep learning model shows more promise for time savings than for increased accuracy. The post-processed model predicts ratios of spine density across genotype conditions similar to counts from our ground-truth Spine Tool dataset using traditional image processing (Figure 5), with an up to 50% reduction in manual correction time.
Improving and Validating Spine Tool
Currently we are optimizing the post-processing steps to ensure that neighboring spines detected by the network are properly segmented and counted. Ultimately, we may be able to detect spines more accurately with deep learning than we can via manual inspection, but that capability is not our primary measure of success. Our goal is to verify that, if a treatment effect can be detected by a human, then it can also be detected by the network.
In the coming months, we will use the new deep learning capabilities of Spine Tool to process images for actual studies. In parallel, we will process the images using our existing workflow and compare the results. If, as we expect, the bar charts produced via the two approaches show comparable average spine density, then we will have confirmed that future studies of spine density can be run reliably with the new tool—and with potentially 50% less manual effort.
Published 2021
References
-
Wu, T. et al. “Complement C3 Is Activated in Human AD Brain and Is Required for Neurodegeneration in Mouse Models of Amyloidosis and Tauopathy.” Cell Reports 28, 2111-2123.e6 (2019).