Phylogenetic Analysis
Build phylogenetic trees from pairwise distances of sequences and determine the evolutionary relationships between organisms. Estimate synonymous and nonsynonymous substitution rates using maximum likelihood. Visualize and edit phylogenetic tree data in the Phylogenetic Tree app.
Apps
| Phylogenetic Tree | Visualize, edit, and explore phylogenetic tree data |
Functions
seqlinkage | Construct phylogenetic tree from pairwise distances |
seqneighjoin | Construct phylogenetic tree using neighbor-joining method |
seqpdist | Calculate pairwise distance between sequences |
phytreeviewer | Visualize, edit, and explore phylogenetic tree data |
dnds | Estimate synonymous and nonsynonymous substitution rates |
dndsml | Estimate synonymous and nonsynonymous substitution rates using maximum likelihood method |
gethmmtree | Retrieve phylogenetic tree data from PFAM database |
seqinsertgaps | Insert gaps into nucleotide or amino acid sequence |
Objects
phytree | Data structure containing phylogenetic tree |
Topics
- Phylogenetic Analysis
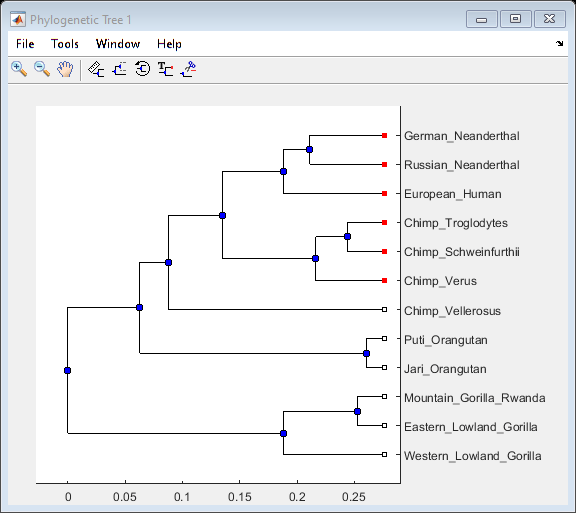
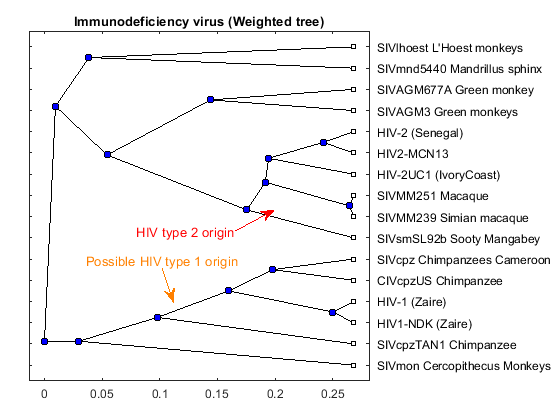
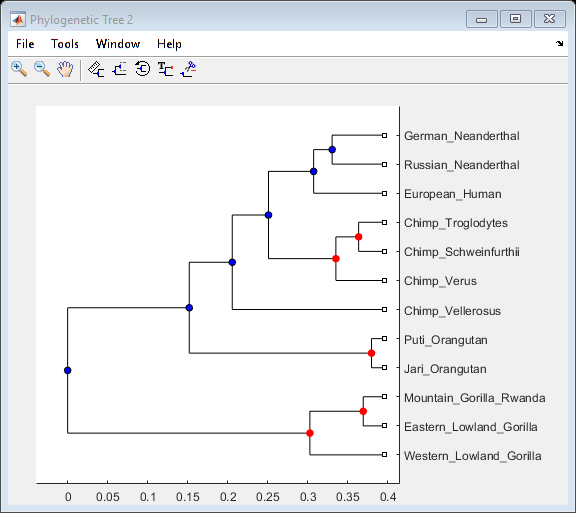
Phylogenetic analysis is the process you use to determine the evolutionary relationships between organisms.
- Using the Phylogenetic Tree App
Description of menu commands and features for creating publishable tree figures.
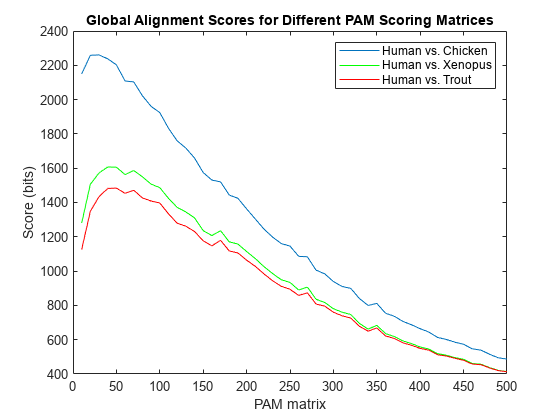
- Sequence Utilities and Statistics
You can manipulate and analyze your sequences to gain a deeper understanding of the physical, chemical, and biological characteristics of your data.