pdbdistplot
Visualize intermolecular distances in Protein Data Bank (PDB) file
Syntax
pdbdistplot(PDBid)
pdbdistplot(PDBid, Distance)
pdbdistplot(___,'Chain',ChainID)
pdbdistplot(___,'Model',ModelNum)
pdbdistplot(___,'Hetero',TF)
Arguments
PDBid | Any of the following:
Note Each structure in the PDB database is represented by a four-character
alphanumeric identifier. For example, |
Distance | Threshold distance in angstroms shown on a spy plot.
Default is |
ChainID | Any of the following:
|
ModelNum | Positive integer specifying which model to consider. Default is 1. |
Description
pdbdistplot displays the distances between
atoms and between residues in a PDB structure.
pdbdistplot( retrieves
the structure specified by PDBid)PDBid from the
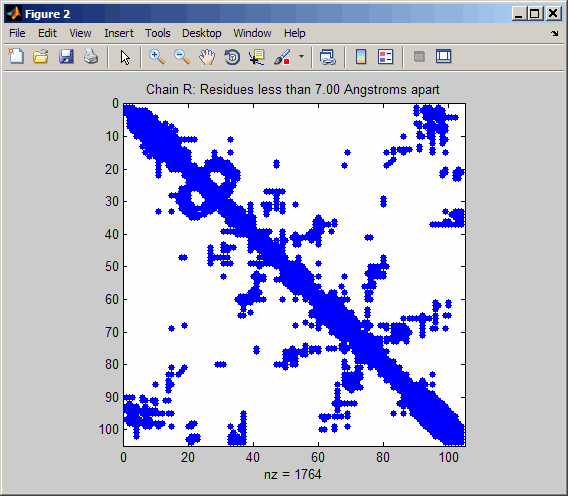
PDB database and creates a heat map showing inter–residue distances
and a spy plot showing the residues where the minimum distances apart
are less than 7 angstroms. If multiple chains are
present in PDBid, separate plots are created.
pdbdistplot( specifies
the threshold distance shown on a spy plot. Default is PDBid, Distance)7.
pdbdistplot(___,'Chain', specifies
the chains to consider. By default, all chains included in the model
are considered.ChainID)
pdbdistplot(___,'Model', specifies
which PDB structural model to consider. Default is 1.ModelNum)
pdbdistplot(___,'Hetero', specifies
whether to include hetero atoms in the plot of residue interactions. TF)TF is
logical, true or false. Default
is false.
Examples
Display a heat map of the inter–residue distances and
a spy plot at 7 angstroms of the protein cytochrome
C from albacore tuna.
pdbdistplot('5CYT');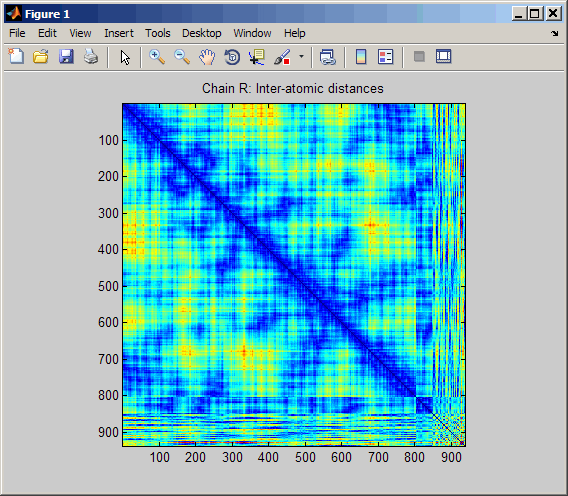
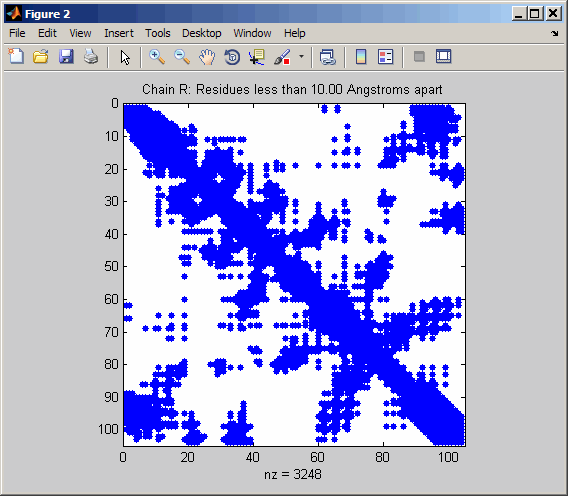
Display a spy plot at 10 angstroms of the
same structure.
pdbdistplot('5CYT',10);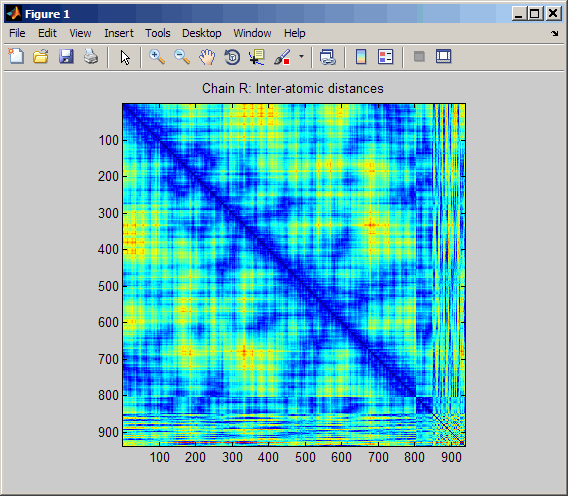
Version History
Introduced before R2006a
See Also
getpdb | pdbread | proteinplot | ramachandran