zonebackadj
Perform background adjustment on Affymetrix microarray probe-level data using zone-based method
Syntax
BackAdjustedData = zonebackadj(Data)
[BackAdjustedData, ZoneStruct]
= zonebackadj(Data)
[BackAdjustedData, ZoneStruct, Background]
= zonebackadj(Data)
... = zonebackadj(Data,
...'NumZones', NumZonesValue, ...)
... = zonebackadj(Data,
...'Percent', PercentValue, ...)
... = zonebackadj(Data,
...'SmoothFactor', SmoothFactorValue, ...)
... = zonebackadj(Data,
...'NoiseFrac', NoiseFracValue, ...)
... = zonebackadj(Data,
...'CDF', CDFValue, ...)
... = zonebackadj(Data,
...'Mask', MaskValue, ...)
... = zonebackadj(Data,
...'Showplot', ShowplotValue, ...)
Input Arguments
Data | Either of the following:
|
NumZonesValue | Scalar or two-element vector that specifies the number of zones
to use in the background adjustment. If a scalar, it must be a square
number. If a two-element vector, the first element specifies the number
of rows and the second element specifies the number of columns in
a nonsquare grid. Default is 16. |
PercentValue | Value that specifies a percentage, P,
such that the lowest P percent of ranked
intensity values from each zone is used to estimate the background
for that zone. Default is 2. |
SmoothFactorValue | Value that specifies the smoothing factor used in the calculation
of the weighted average of the contributions of each zone to the background
of a point. Default is 100. |
NoiseFracValue | Value that specifies the noise fraction, NF,
such that the background-adjusted value is given by max((Intensity
- WeightedBackground), NF*LocalNoiseEstimate). Default is 0.5. |
CDFValue | Either of the following:
|
MaskValue | Logical vector that specifies which cells to mask and not use
in the background estimates. In the vector, 0 = not masked and 1
= masked. Defaults are the values in the Masked column
of the Probes field of the CEL file. |
ShowplotValue | Controls the plotting of an image of the background estimates.
Choices are |
Output Arguments
BackAdjustedData | Matrix or cell array of vectors containing background-adjusted probe intensity values. |
ZoneStruct | MATLAB structure containing the centers of the zones used to perform the background adjustment and the estimates of the background values at the center of each zone. |
Background | Matrix or cell array of vectors containing the estimated background values for each probe. |
Description
BackAdjustedData = zonebackadj(Data)Data, which
contains probe intensities from Affymetrix CEL files. Details of the background adjustment are described in Statistical Algorithms Description Document.
[ also returns
a structure containing the centers of the zones used to perform the
background adjustment and the estimates of the background values at
the center of each zone. BackAdjustedData, ZoneStruct]
= zonebackadj(Data)
[ also returns
a matrix or cell array of vectors containing the estimated background
values for each probe. BackAdjustedData, ZoneStruct, Background]
= zonebackadj(Data)
... = zonebackadj( calls Data,
...'PropertyName', PropertyValue,
...)zonebackadj with optional
properties that use property name/property value pairs. You can specify
one or more properties in any order. Each PropertyName must
be enclosed in single quotation marks and is case insensitive. These
property name/property value pairs are as follows:
... = zonebackadj( specifies
the number of zones to use in the background adjustment. Data,
...'NumZones', NumZonesValue, ...)NumZonesValue can
be either a scalar that is a square number or a two-element array
in which the first element specifies the number of rows and the second
element specifies the number of columns in a nonsquare grid. Default
is 16.
... = zonebackadj( specifies
a percentage, Data,
...'Percent', PercentValue, ...)P, such that the lowest P percent
of ranked intensity values from each zone is used to estimate the
background for that zone. Default is 2.
... = zonebackadj( specifies
the smoothing factor used in the calculation of the weighted average
of the contributions of each zone to the background of a point, thus
providing a smooth transition between zones. Default is Data,
...'SmoothFactor', SmoothFactorValue, ...)100.
... = zonebackadj( specifies
the noise fraction, such that the background-adjusted value is given
by Data,
...'NoiseFrac', NoiseFracValue, ...)max((Intensity - WeightedBackground), NF*LocalNoiseEstimate),
where NF is NoiseFracValue.
Default is 0.5.
... = zonebackadj( specifies
an Affymetrix CDF library file or structure, which specifies
control cells, which are not used in the background estimates.Data,
...'CDF', CDFValue, ...)
... = zonebackadj( specifies
a logical vector of that specifies which cells to mask and not use
in the background estimates. In the vector, Data,
...'Mask', MaskValue, ...)0 = not masked and 1
= masked. Defaults are the values in the Masked column
of the Probes field of the CEL file.
... = zonebackadj( plots
an image of the background estimates. Choices are Data,
...'Showplot', ShowplotValue, ...)true or false (default).
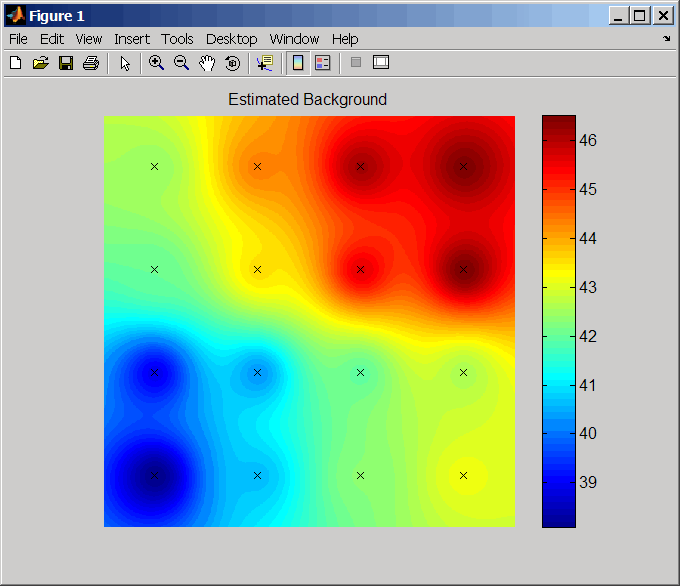
Examples
References
[1] Statistical Algorithms Description Document, https://assets.thermofisher.com/TFS-Assets/LSG/brochures/sadd_whitepaper.pdf
Version History
Introduced in R2007b
See Also
affyinvarsetnorm | affyread | celintensityread | gcrma | gcrmabackadj | probelibraryinfo | probesetlookup | probesetvalues | quantilenorm | rmabackadj | rmasummary
