View and Align Multiple Sequences
Overview of the Sequence Alignment App
The Sequence Alignment app integrates many sequence and multiple alignment functions in the toolbox. Instead of entering commands in the MATLAB® Command Window, you can use this app to visually inspect a multiple alignment and make manual adjustments.
Visualize Multiple Sequence Alignment
Read a multiple sequence alignment file of the gag polyprotein for several HIV strains.
gagaa = multialignread('aagag.aln')View the aligned sequences in the Sequence Alignment app.
seqalignviewer(gagaa);

Adjust Sequence Alignments Manually
Algorithms for aligning multiple sequences do not always produce an optimal result. By visually inspecting the alignment, you can identify areas whose alignment can be improved by a manual adjustment.
To better visualize the sequence alignments, you can zoom in by selecting Display > Zoom in. Select this option multiple times until you achieve the zoom level you want.
Identify an area where you could improve the alignment.
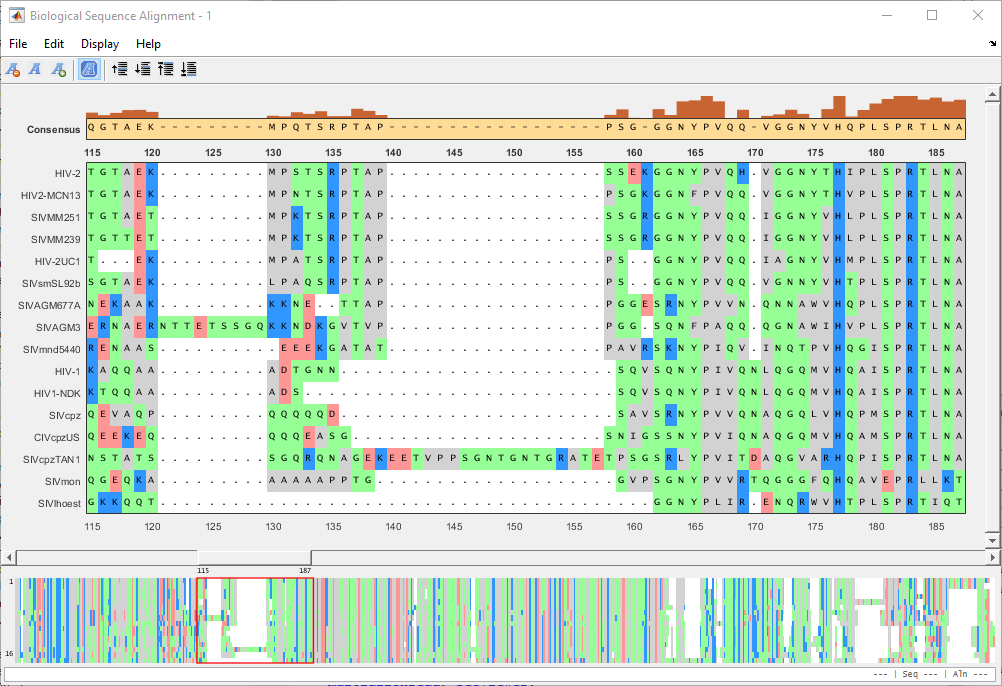
Click a letter or a region. The selected region is the center block. You can then drag the sequence(s) to the left or right of the center block.
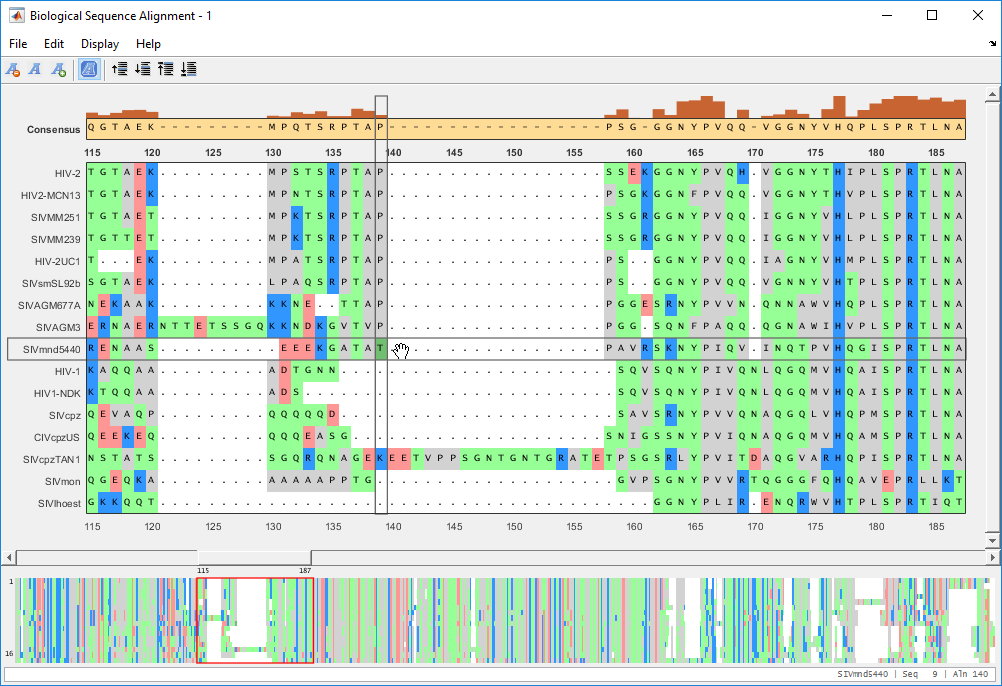
To move a single letter (T in this example), click and drag the letter T (center block) to the right to insert a gap.
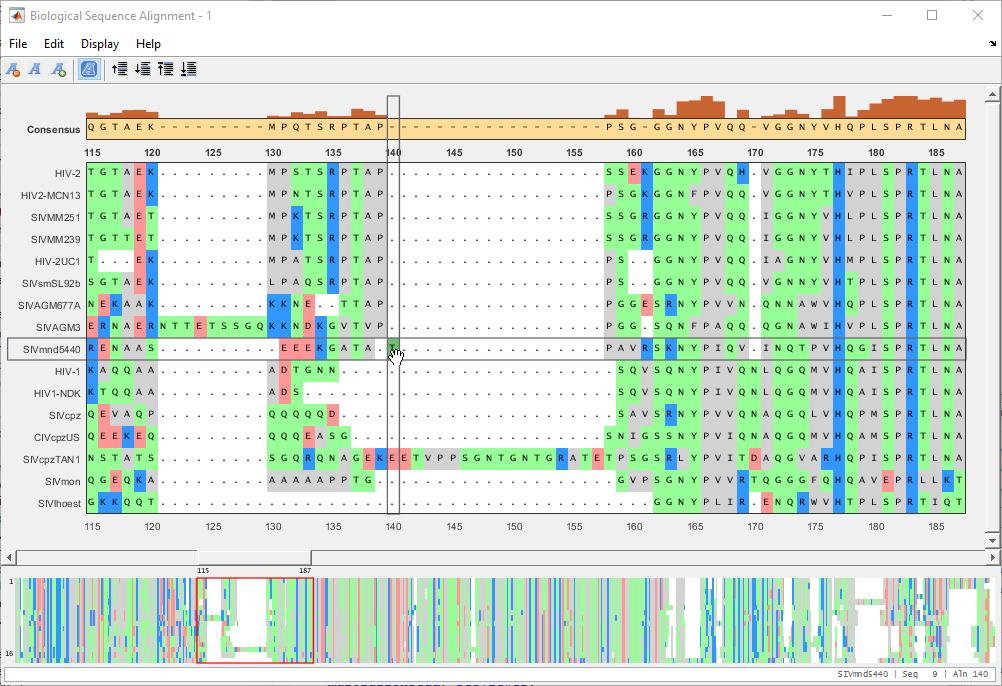
Close the gap by dragging the letter back to the left.
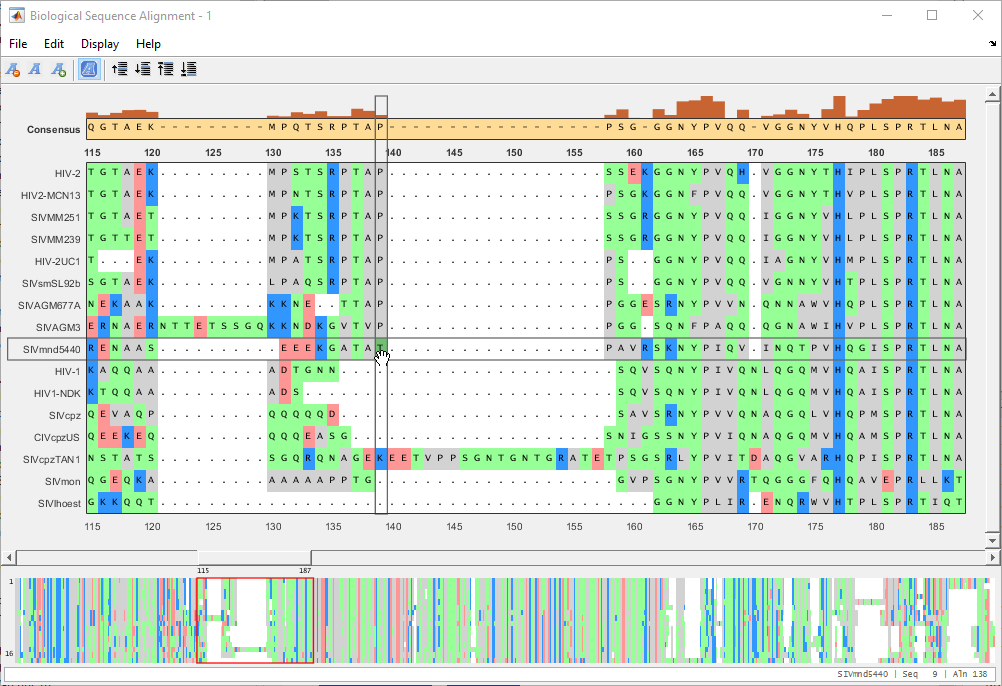
You can also move multiple residues (a subsequence). Suppose you want to move a subsequence to available gaps. First select the gap region that you want to fill in.

Drag the subsequence(s) from the right or left of the gap region into the gap area.

Suppose you want to remove one or more of the aligned sequences. First select the sequence(s) to be removed. Then select Edit > Delete Sequences.
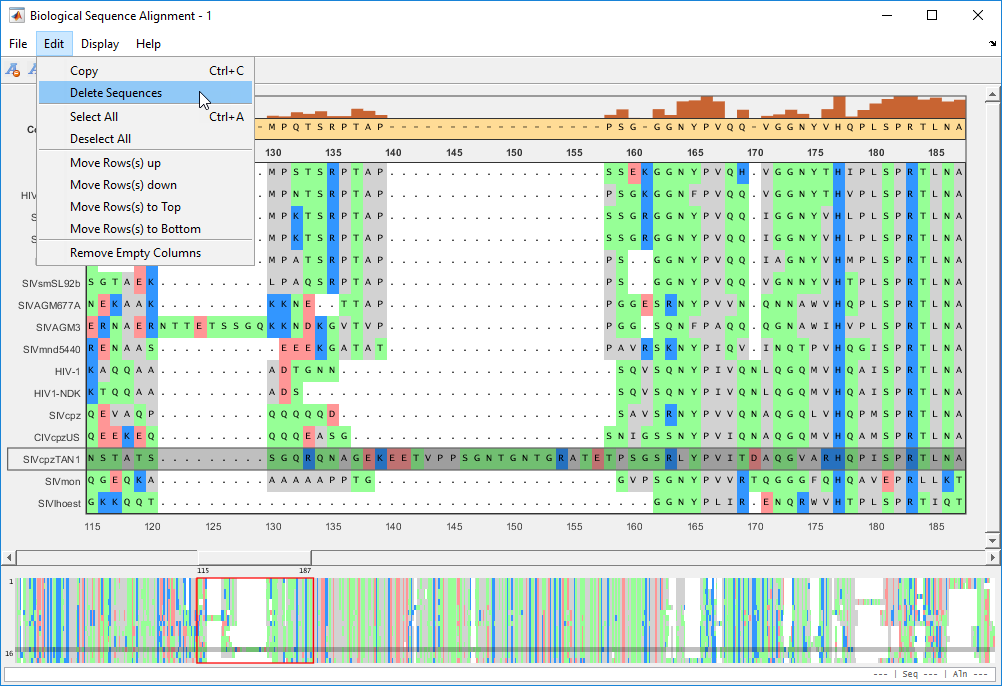
Remove empty columns by selecting Edit > Remove Empty Columns.
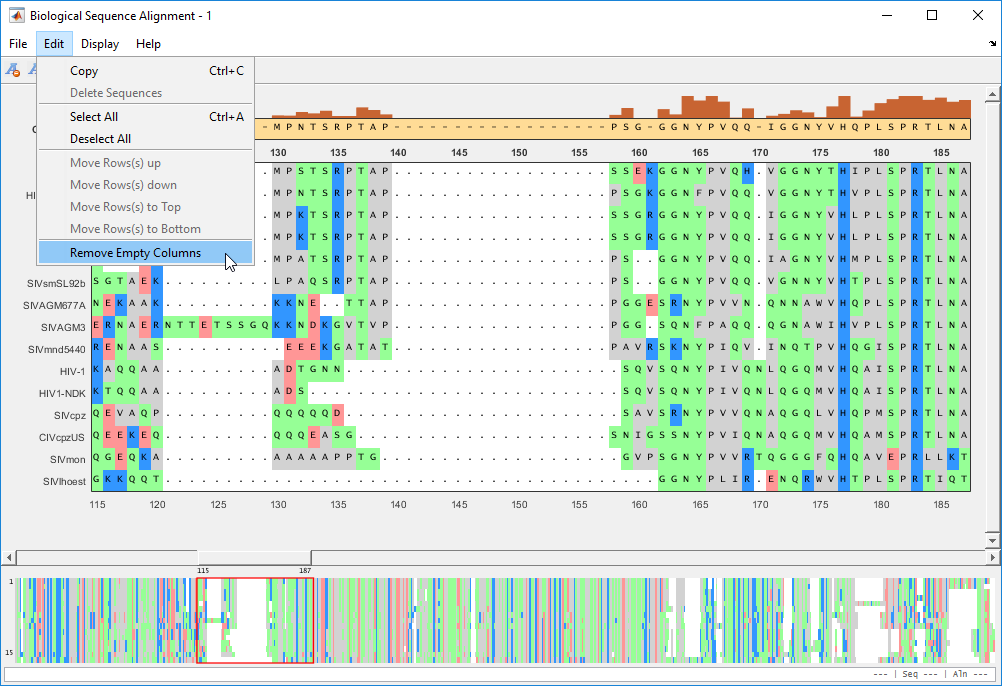
After the edit, you can export the aligned sequences or consensus sequence to a FASTA file or MATLAB Workspace from the File menu.
Rearrange Rows
You can move the rows (sequences) up or down by one row. You can also move selected rows to the top or bottom of the list.
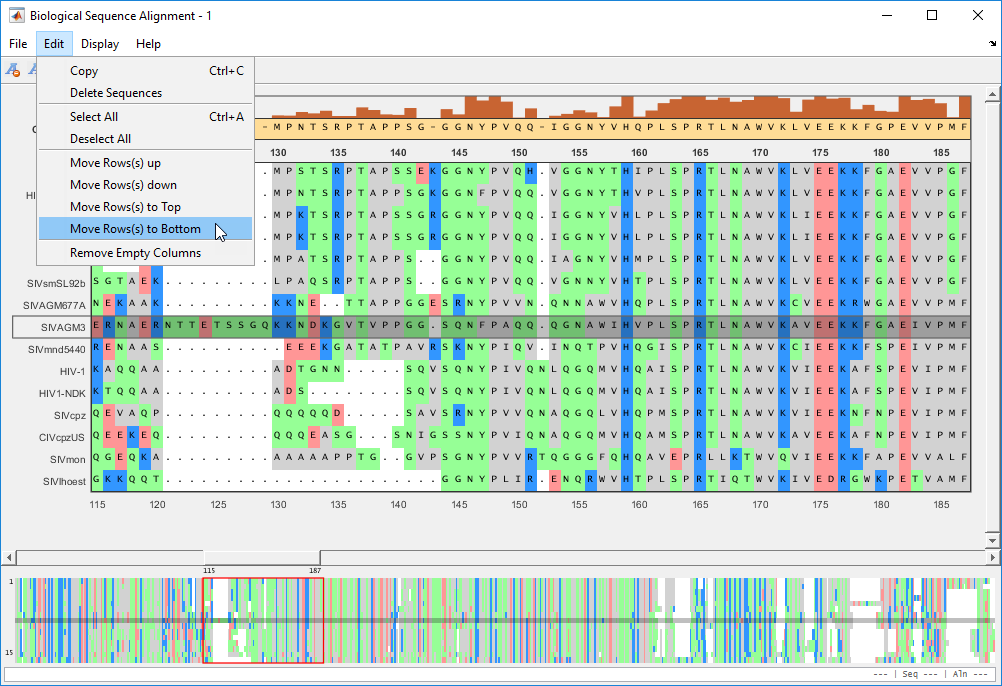
The selected sequence moves to the bottom of the list.

Generate Phylogenetic Tree from Aligned Sequences
You can generate a phylogenetic tree using the aligned sequences from within the app. You can select a subset of sequences or use all the sequences to generate a tree.
Select Display > View Tree > Selected... to generate a tree from selected sequences.

A phylogenetic tree for the sequences is displayed in the Phylogenetic Tree app. For details on the app, see Using the Phylogenetic Tree App.

See Also
seqalignviewer | Sequence Alignment | Genomics
Viewer