Estimate Lung Displacement Field During Breathing Using Deformable Image Registration
This example uses deformable image registration to estimate the displacement field between two CT scans acquired during inhalation and exhalation for the same patient.
Deformable image registration aligns 3-D volumes while accounting for non-rigid motion, such as expanding and contracting lung tissue. Deformable registration of CT scans acquired during inhalation and exhalation can map the displacement of lung tissue during the two phases of breathing. Quantifying lung motion is useful for applications such as radiotherapy planning for lung cancer, because precisely targeting treatment can help to avoid damaging healthy tissue.
Download Data
This example uses a subset of data from one subject in a data set containing CT and PET images [1][2]. The subset includes one CT scan acquired during inhalation and one CT scan acquired during exhalation. The size of the subset of data is approximately 137 MB. Run this code to download the data set from the MathWorks® website, and then unzip the folder.
zipFile = matlab.internal.examples.downloadSupportFile("medical", ... "CT-PET-Ventilation-Imaging/Chest-Ventilation-Imaging.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
After you run the code to download the data, the dataFolder folder contains the downloaded and unzipped data. Each scan consists of a directory of DICOM files.
dataFolder = fullfile(filepath,"Chest-Ventilation-Imaging");Import Data
Read the CT volume acquired at 80% of maximum inhalation as a medicalVolume object.
medVolInsp = medicalVolume(fullfile(dataFolder,"5.000000-Thorax Insp 2.0 B70f-84301"));Read the CT volume acquired at peak exhalation as a medicalVolume object.
medVolExsp = medicalVolume(fullfile(dataFolder,"7.000000-Thorax Exp 2.0 B70f-98860"));Display Unregistered Volumes
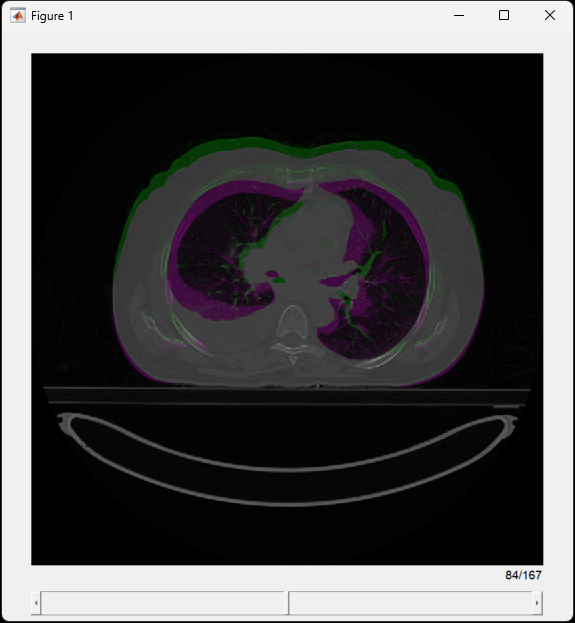
To visualize differences within the lungs, create a falsecolor overlay of the CT volumes by using the displayFusedSlices helper function, defined at the end of this example.
displayFusedSlices(medVolInsp,medVolExsp)
This image shows the middle transverse slice. Purple and green indicate where the images are different. In the inhalation image, the anterior wall of the abdomen is extended and the lung cavity is larger as compared to the exhalation image.
Calculate Deformable Registration
Register the CT volume at inhalation to the CT volume at exhalation by using the imregdeform function. The registration can take several minutes. The name-value arguments specified in this example have been determined for this data set by using trial and error.
moving = medVolInsp.Voxels;
fixed = medVolExsp.Voxels;
[dispField,reg] = imregdeform(moving,fixed,PixelResolution=medVolInsp.VoxelSpacing, ...
NumPyramidLevels=6,GridRegularization=0.01);--------------------------- Pyramid Level = 6 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 1.62481 39852990.50 14.24 5685.85944
2 1.62183 38583968.52 37.41 3963.36302
3 1.62707 36918008.24 142.41 6034.82919
4 1.61063 35500788.44 55.79 5876.61545
5 1.62511 34369435.01 73.64 8360.93571
6 1.60027 33748504.42 30.29 4118.84221
7 1.58207 33258484.20 37.79 5999.56215
8 1.57513 32963564.36 23.75 5731.53510
9 1.58490 32651994.46 63.44 9566.42583
10 1.58375 32388652.65 27.98 4869.31721
11 1.57107 32194121.93 29.89 2920.65547
12 1.57197 32011187.89 78.36 10958.16473
13 1.57276 31927547.62 46.60 8055.35878
14 1.56669 31772759.29 21.18 4067.68143
15 1.55234 31691226.84 41.83 3398.35486
16 1.54753 31562509.85 24.18 3955.47316
--------------------------- Pyramid Level = 5 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 0.91176 42907652.78 7.65 7088.24860
2 0.89018 41640846.07 64.12 4231.25487
3 0.88098 41268677.48 23.56 2308.48118
4 0.88108 40420137.71 69.98 3017.36869
5 0.88136 39708273.80 94.18 3004.63424
6 0.86391 39287029.08 53.18 2836.02841
7 0.85912 38752913.15 81.04 2839.52290
8 0.85921 38451159.94 72.15 3121.08355
9 0.85602 38316538.90 91.19 2466.11900
10 0.84566 38002862.66 83.00 2364.25555
11 0.83822 37861145.02 59.98 2403.32790
12 0.84172 37756426.21 75.65 2078.77143
13 0.83826 37595486.37 61.51 1910.89786
14 0.83233 37531809.31 79.86 2722.11701
15 0.82924 37400463.88 15.97 2709.45434
16 0.82911 37387959.64 0.00 2707.00988
--------------------------- Pyramid Level = 4 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 0.47753 48456268.43 12.98 1654.79835
2 0.46451 46594384.96 105.95 4570.51898
3 0.46212 45700690.01 71.53 1379.37637
4 0.44874 45031332.42 63.87 3161.53525
5 0.44395 44187748.79 122.64 2451.07500
6 0.43571 43665724.89 79.15 1377.44182
7 0.42269 43199527.35 85.00 1951.32573
8 0.41817 42970615.77 105.08 2158.87794
9 0.41616 42735146.97 97.18 1575.88025
10 0.40931 42474966.48 105.66 1777.79492
11 0.40414 42458729.40 35.24 1350.66373
12 0.41312 42283387.80 143.35 2041.06972
13 0.41310 42225888.11 0.00 2042.91928
--------------------------- Pyramid Level = 3 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 0.22783 49311583.84 143.88 2267.06525
2 0.21111 48178102.47 218.63 1339.66693
3 0.19817 47228202.76 95.12 1379.83890
4 0.19378 45851874.77 179.58 1304.66621
5 0.19193 44838936.48 130.51 978.18151
6 0.17652 44326943.08 97.03 1180.80566
7 0.17294 43924479.14 110.20 1313.57415
8 0.17548 43496462.62 181.92 1116.91897
9 0.17084 43300202.82 149.20 1236.50319
10 0.16356 43010008.95 118.90 802.53488
11 0.16478 42881600.54 123.67 1049.02394
12 0.16258 42724378.49 122.59 674.90351
13 0.15803 42607153.59 150.11 1110.24069
14 0.15519 42576294.01 36.96 799.21994
15 0.15154 42483035.81 40.07 517.67688
16 0.14836 42451062.84 55.86 518.14514
--------------------------- Pyramid Level = 2 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 0.08422 47083777.99 28.30 397.92074
2 0.08635 45453138.20 252.18 1016.62827
3 0.08040 44767616.45 95.85 517.63867
4 0.07262 44189019.22 114.66 408.32415
5 0.07737 43623129.27 242.67 445.97621
6 0.07611 43562484.48 63.48 450.35100
7 0.07232 43423602.26 309.42 635.36467
8 0.06811 43178330.49 182.88 573.87771
9 0.06667 43087998.21 134.52 624.74004
10 0.07124 42857329.40 221.50 520.82692
11 0.06969 42804593.41 42.17 406.47788
12 0.06762 42787115.63 265.57 723.87049
--------------------------- Pyramid Level = 1 --------------------------------
Normalized Function local Closeness to
Iteration rmse minima Step-size optimal solution
1 0.04009 50884600.63 246.21 517.74695
2 0.04005 50136965.51 192.07 440.97480
3 0.03844 49281978.34 178.08 633.73365
4 0.03850 48691759.02 243.37 436.44544
5 0.03779 48426211.65 248.42 449.98833
6 0.03589 48059453.05 240.01 485.05931
7 0.03509 47866744.43 71.46 558.09062
8 0.03444 47731975.74 132.79 322.34538
9 0.03444 47597203.08 127.25 327.93428
10 0.03442 47538280.88 97.17 333.62361
11 0.03436 47513235.40 99.72 322.89874
12 0.03428 47336573.73 21.56 363.61216
Create a new medicalVolume object that contains the registered image data and the spatial referencing information for the fixed exhale volume.
medVolReg = medicalVolume(reg,medVolExsp.VolumeGeometry);
Display Registered Volumes
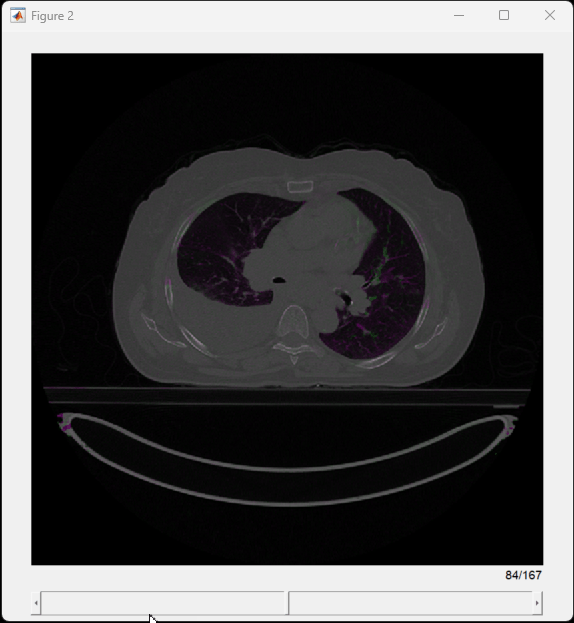
To visually assess the registration, create a falsecolor overlay of the fixed volume and transformed moving volume by using the displayFusedSlices helper function.
displayFusedSlices(medVolReg,medVolExsp)
Display Displacement Field
In addition to the registered image data, the imregdeform function returns an m-by-n-p-by-3 displacement field. The first three dimensions correspond to row, column, and slice indices of the registered volume, and the fourth dimension contains the displacements in the xyz-directions, in pixels. For example, the value dispfield(1,1,1,:) returns a 3-element vector containing the xyz-displacements at the voxel location (1,1,1).
whos dispfieldConvert the displacements from pixel units to millimeters by multiplying each of the x-, y- and z-displacements by the corresponding voxel spacing value.
dispFieldMM = dispField.*reshape(medVolInsp.VoxelSpacing,[1 1 1 3]);
Calculate the magnitude of the displacement by using the vecnorm function. Specify the p input as 2 to calculate the Euclidean distance, and the dimension along which to calculate the norm as 4.
dispMag = vecnorm(dispFieldMM,2,4);
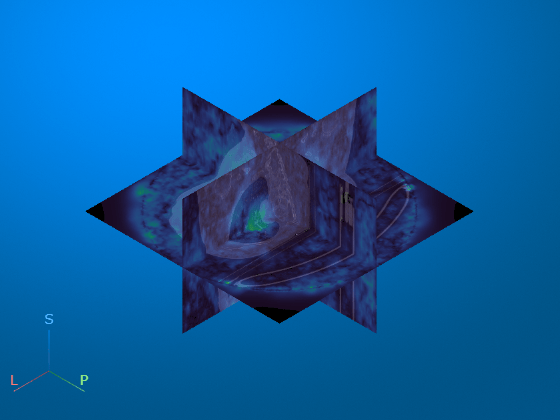
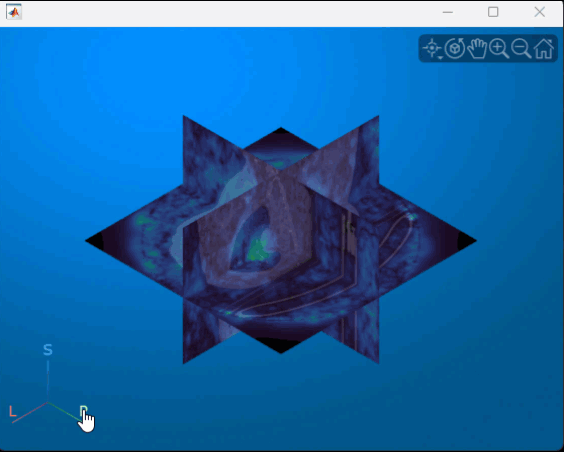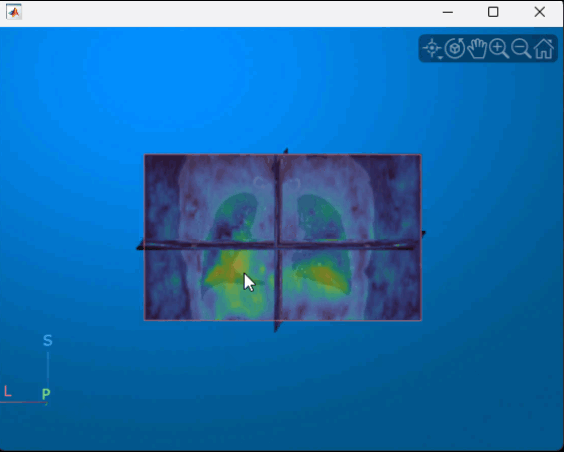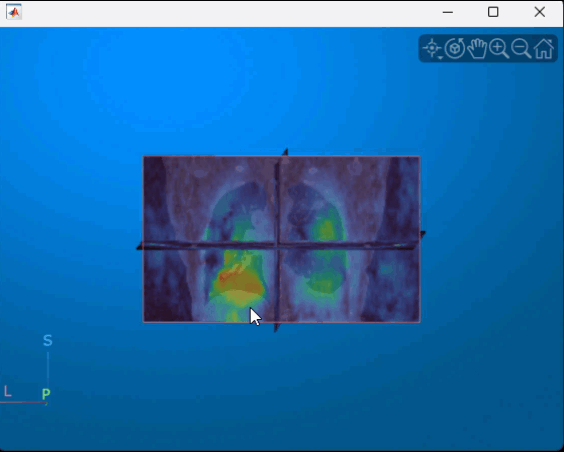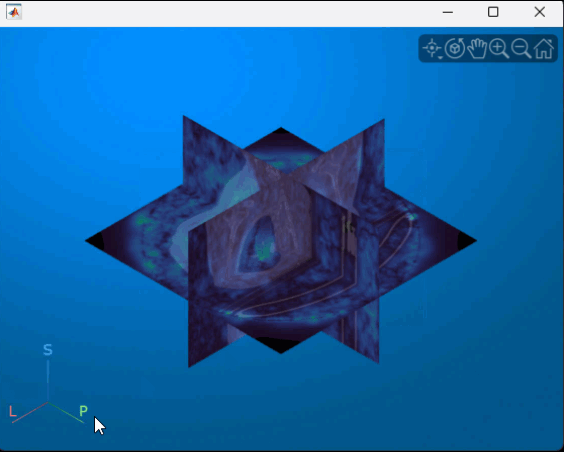
Display the displacement magnitude as an overlay over the original inhale CT volume. The cool heatmap colors indicate smaller displacements, and the warm heatmap regions indicate larger displacements. As expected, the largest respiratory motion occurs within the bottom of the lungs, near the diaphragm. Regions outside the lungs have minimal motion, as the patient did not move or change position within the scanner other than to breathe.
volshow(medVolInsp, ... RenderingStyle="SlicePlanes", ... OverlayData=dispMag, ... OverlayDisplayRangeMode="data-range", ... OverlayColormap=turbo(256), ... OverlayAlphamap=0.5);
You can interact with the volume display to explore the displacement data, as shown in this animation. To view the coronal plane, click the P axes display indicator. Drag the cursor to rotate the volume slightly out of plane. To scroll through the coronal slices, pause on the coronal slice until it highlights red, then drag it along the anterior/posterior axis. To rotate the volume, click anywhere in the viewer and drag.
Supporting Function
The displayFusedSlices helper function displays a fused overlay of transverse slices of two medicalVolume objects by performing these steps:
To apply uniform scaling to all slices, rescale the voxel intensities to the range [0,1].
Loop through the volumes in the third dimension. At each step, extract the current slice from each volume, and create a fused falsecolor overlay by using
imfuse. Specify theScalingname-value argument as"none"to avoid rescaling the intensity of each slice separately. Add the fused slice to the arrayfused.To display the fused image as an RGB volume, reorder the dimensions of the fused image volume so that
fusedPermutedis an m-by-n-by-p-by-3 array.Create a new
medicalVolumeobject that contains the fused image data and the same spatial referencing as the original volumes.Display the slices of
medVolFusedin a scrollable figure window.
function displayFusedSlices(medVol1,medVol2) medVol1Rescale = medVol1; medVol1Rescale.Voxels = rescale(medVol1.Voxels); medVol2Rescale = medVol2; medVol2Rescale.Voxels = rescale(medVol2.Voxels); for i = 1:size(medVol1Rescale.Voxels,3) slice1 = medVol1Rescale.Voxels(:,:,i); slice2 = medVol2Rescale.Voxels(:,:,i); fused(:,:,:,i) = imfuse(slice1,slice2,method="falsecolor",Scaling="none"); end fusedPermuted = permute(fused,[1 2 4 3]); medVolFused = medicalVolume(fusedPermuted,medVol1.VolumeGeometry); sliceViewer(medVolFused) end
References
[1] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. “CT Ventilation as a Functional Imaging Modality for Lung Cancer Radiotherapy (CT-vs-PET-Ventilation-Imaging).” The Cancer Imaging Archive, 2022. https://doi.org/10.7937/3PPX-7S22.
[2] Eslick, Enid M., John Kipritidis, Denis Gradinscak, Mark J. Stevens, Dale L. Bailey, Benjamin Harris, Jeremy T. Booth, and Paul J. Keall. “CT Ventilation Imaging Derived from Breath Hold CT Exhibits Good Regional Accuracy with Galligas PET.” Radiotherapy and Oncology 127, no. 2 (May 2018): 267–73. https://doi.org/10.1016/j.radonc.2017.12.010.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
See Also
imregdeform | sliceViewer | volshow | imfuse