Create and Simulate a Model with a Custom Function
This example shows how to create a custom function and incorporate it in model simulation.
Overview
Prerequisites for the Example
This example assumes that you have a working knowledge of:
MATLAB® app
Creating and saving MATLAB programs
About the Example Model
This example uses the model described in Model of the Yeast Heterotrimeric G Protein Cycle.
This table shows the reactions used to model the G protein cycle and the corresponding rate parameters (rate constants) for each reaction. For reversible reactions, the forward rate parameter is listed first.
| No. | Name | Reaction1 | Rate Parameters |
|---|---|---|---|
| 1 | Receptor-ligand interaction | L + R <-> RL | kRL, kRLm |
| 2 | Heterotrimeric G protein formation | Gd + Gbg -> G | kG1 |
| 3 | G protein activation | RL + G -> Ga + Gbg + RL | kGa |
| 4 | Receptor synthesis and degradation | R <-> null | kRdo, kRs |
| 5 | Receptor-ligand degradation | RL -> null | kRD1 |
| 6 | G protein inactivation | Ga -> Gd | kGd |
| 1 Legend of species: L = ligand (alpha factor), R = alpha-factor receptor, Gd = inactive G-alpha-GDP, Gbg = free levels of G-beta:G-gamma complex, G = inactive Gbg:Gd complex, Ga = active G-alpha-GTP | |||
Assumptions of the Example
This example assumes that:
An inhibitor (
Inhibspecies) slows the inactivation of the active G protein (reaction 6 above,Ga –> Gd).The variation in the amount of inhibitor (
Inhibspecies) is defined in a custom function,inhibvalex.The inhibitor (
Inhibspecies) affects the reaction by changing the amount of rate parameterkGd.
About the Example
This example shows how to create and call a custom function in a SimBiology® expression. Specifically, it shows how to use a custom function in a rule expression.
About Using Custom Functions in SimBiology Expressions
You can use custom functions in:
Reaction rate expressions (
ReactionRateproperty)Rule expressions (
Ruleproperty)Event expressions (
EventFcnsproperty orTriggerproperty)
The requirements for using custom functions in SimBiology expressions are:
Create a custom function. For more information, see
function.Change the current folder to the folder containing your custom MATLAB file. Do this by using the
cdcommand or by using the Current Folder field in the MATLAB desktop toolbar. Alternatively, add the folder containing your file to the search path. Do this by using theaddpathcommand or see Change Folders on Search Path.Call the custom function in a SimBiology reaction, rule, or event expression.
Tip
If your rule or reaction rate expression is not continuous and differentiable, see Using Events to Address Discontinuities in Rule and Reaction Rate Expressions before simulating your model.
Create a Custom Function
The following procedure creates a custom function,
inhibvalex, which lets you specify how the inhibitor
amount changes over time. The inputs are time, the initial amount of inhibitor,
and a parameter that governs the amount of inhibitor. The output of the function
is the amount of inhibitor.
In the MATLAB desktop, select File > New > Script, to open the MATLAB Editor.
Copy and paste the following function declaration:
% inhibvalex.m function Cp = inhibvalex(t, Cpo, kel) % This function takes the input arguments t, Cpo, and kel % and returns the value of the inhibitor Cp. % You can later specify the input arguments in a % SimBiology rule expression. % For example in the rule expression, specify: % t as time (a keyword recognized as simulation time), % Cpo as a parameter that represents the initial amount of inhibitor, % and kel as a parameter that governs the amount of inhibitor. if t < 400 Cp = Cpo*exp(-kel*(t)); else Cp = Cpo*exp(-kel*(t-400)); end
Save the file (name the file inhibvalex.m) in a
directory that is on the MATLAB search path, or to a directory that you can access.
If the location of the file is not on the MATLAB search path, change the working directory to the file location.
Load the Example Model
Load the gprotein example project, which includes the variable
m1, a model object:
sbioloadproject gproteinThe m1 model object appears in the MATLAB Workspace.
Add the Custom Function to the Example Model
The following procedure creates a rule expression that calls
the custom function, inhibvalex, and specifies the three
input values to this function.
Add a repeated assignment rule to the model that specifies the three input
values to the custom function, inhibvalex:
rule1 = addrule(m1, 'Inhib = inhibvalex(time, Cpo, Kel)',... 'repeatedAssignment');
The time input is a SimBiology keyword recognized as simulation time
Create the two parameters used by the rule1 rule and
assign values to them:
p1 = addparameter(m1, 'Cpo', 250); p2 = addparameter(m1, 'Kel', 0.01);
Create the species used by the rule1 rule:
s1 = addspecies(m1.Compartments, 'Inhib');Define a Rule to Change Parameter Value
The value of rate parameter kGd is
affected by the amount of inhibitor present in the system. Add a rule to the
model to describe this action, but first change the
ConstantValue property of the parameter
kGd so that it can be varied by a rule.
Change the ConstantValue property of the
kGd parameter to false.
p3 = sbioselect(m1, 'Type', 'parameter', 'Name', 'kGd'); p3.ConstantValue = false;
Add a repeated assignment rule to the model to define how the
kGd parameter is affected by the
Inhib species.
rule2 = addrule(m1, 'kGd = 1/Inhib', 'repeatedAssignment');
Add an Event to Reset the Solver at a Discontinuity and Simulate the Model
The custom function, inhibvalex,
introduces a discontinuity in the model when time = 400. To ensure accurate
simulation results, add an event to the model to reset the solver at the time of
the discontinuity. Set the event to trigger at the time of the discontinuity
(time = 400). The event does not need to modify the model, so create an event
function that multiplies a species value by 1.
addevent(m1, 'time>=400', 'G=1*G');
Configure the simulation settings (configset object)
for the m1 model object to log all states during the
simulation.
cs = getconfigset(m1);
cs.RuntimeOptions.StatesToLog = 'all';
Simulate the model.
simDataObj = sbiosimulate(m1);
Plot the results.
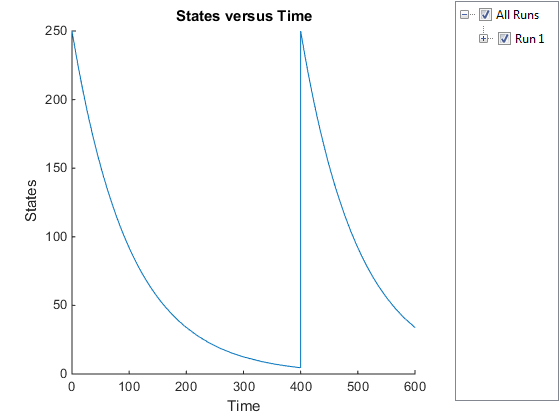
sbioplot(simDataObj);
The plot does not show the species of interest due to the wide range in species amounts/concentrations.
Plot only the species of interest. Ga.
GaSimDataObj = selectbyname(simDataObj,'Ga');
sbioplot(GaSimDataObj);
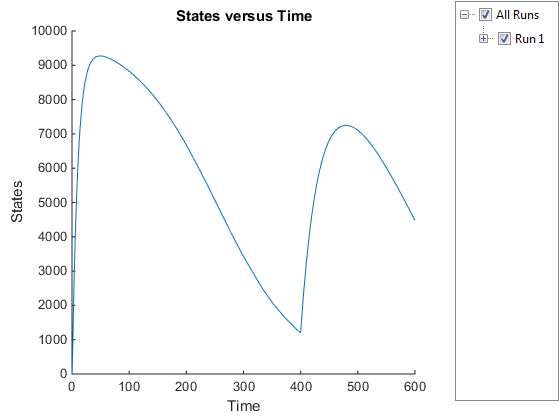
Notice the change in the profile of species Ga at time
= 400 seconds (simulation time). This is the time when
the inhibitor amount is changed to reflect the re-addition of inhibitor to
the model.
Plot only the inhibitor (Inhib species).
InhibSimDataObj = selectbyname(simDataObj,'Inhib');
sbioplot(InhibSimDataObj)