Wavelet Multiscale Principal Components Analysis
This example demonstrates the features of multiscale principal components analysis (PCA) provided in Wavelet Toolbox™.
The aim of multiscale PCA is to reconstruct, starting from a multivariate signal and using a simple representation at each resolution level, a simplified multivariate signal. The multiscale principal components analysis generalizes the normal PCA of a multivariate signal represented as a matrix by performing a PCA on the matrices of details of different levels simultaneously. A PCA is also performed on the coarser approximation coefficients matrix in the wavelet domain as well as on the final reconstructed matrix. By selecting the numbers of retained principal components, interesting simplified signals can be reconstructed.
Import Data
Load the multivariate dataset.
load ex4mwden
whosName Size Bytes Class Attributes covar 4x4 128 double x 1024x4 32768 double x_orig 1024x4 32768 double
Usually, only the matrix of data x is available. Here, we also have the true noise covariance matrix covar and the original signals x_orig. These signals are noisy versions of simple combinations of the two original signals. The first signal is "Blocks", which is irregular, and the second one is "HeavySine", which is regular, except around time 750. The other two signals are the sum and the difference of the two original signals, respectively. Multivariate Gaussian white noise exhibiting strong spatial correlation is added to the resulting four signals, which produces the observed data stored in x.
Plot the original signals and the signals with additive noise.
tiledlayout(4,2) for i = 1:4 nexttile plot(x_orig(:,i)) axis tight title("Clean Signal "+num2str(i)) nexttile plot(x(:,i)) axis tight title("Noisy Signal "+num2str(i)) end

Simple Multiscale PCA
Wavelet multiscale PCA combines noncentered PCA on approximations and details in the wavelet domain and a final PCA. At each level, the most significant principal components are selected.
Use the wmspca function to perform a multiscale PCA of the noisy signals. Specify the sym4 wavelet and a decomposition down to level 5. Specify the wavelet parameters. Use Kramer's rule to automatically select the number of retained principal components. Kaiser's rule retains the components associated with eigenvalues exceeding the mean of all eigenvalues.
wname = "sym4"; level = 5; npc = "kais"; [x_sim,qual,npc] = wmspca(x,level,wname,npc);
Display the original noisy and simplified signals.
tiledlayout(4,2) for i = 1:4 nexttile plot(x(:,i)) set(gca,'xtick',[]) axis tight title("Noisy Signal "+num2str(i)) nexttile plot(x_sim(:,i)) set(gca,'xtick',[]) axis tight title("Simplified Signal "+num2str(i)) end

The results from a compression perspective are good. Inspect the quality of the reconstructions. The percentages reflecting the quality of column reconstructions given by the relative mean square errors are close to 100%.
qual
qual = 1×4
98.0545 93.2807 97.1172 98.8603
Improve Results
You can improve the results by suppressing noise, because the details at levels 1 to 3 are composed essentially of noise with small contributions from the signal. Removing the noise leads to a crude, but large, denoising effect.
The output argument npc contains the numbers of retained principal components selected by Kaiser's rule.
npc
npc = 1×7
1 1 1 1 1 2 2
For from 1 to 5, is the number of retained noncentered principal components (PCs) for details at level d. The number of retained noncentered PCs for approximations at level 5 is npc(6), and npc(7) is the number of retained PCs for final PCA after wavelet reconstruction. As expected, the rule keeps two principal components, both for the PCA approximations and the final PCA, but one principal component is kept for details at each level.
Suppress the details at levels 1 to 3 by first setting the corresponding elements of npc to 0. Then perform a second multiscale PCA using the modified npc as input.
npc(1:3) = [0 0 0]; [x_sim,qual,npc] = wmspca(x,level,wname,npc);
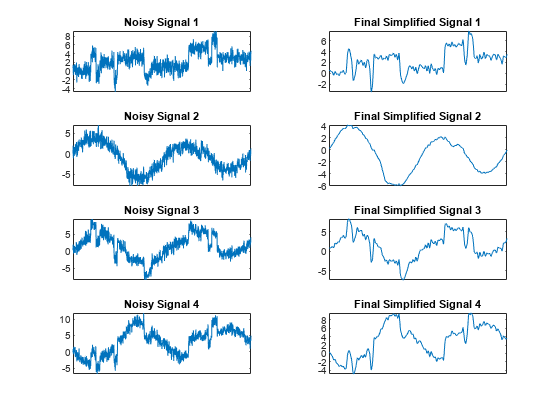
Display the original noisy and final simplified signals. By suppressing the details at levels 1 to 3, the results are improved.
tiledlayout(4,2) for i = 1:4 nexttile plot(x(:,i)) set(gca,'xtick',[]) axis tight title("Noisy Signal "+num2str(i)) nexttile plot(x_sim(:,i)) set(gca,'xtick',[]) axis tight title("Final Simplified Signal "+num2str(i)) end
References
[1] Aminghafari, Mina, Nathalie Cheze, and Jean-Michel Poggi. “Multivariate Denoising Using Wavelets and Principal Component Analysis.” Computational Statistics & Data Analysis 50, no. 9 (May 2006): 2381–98. https://doi.org/10.1016/j.csda.2004.12.010.