使用 K 均值聚类实现基于颜色的分割
此示例说明如何使用 K 均值聚类自动分割颜色。
聚类是一种分离目标组的方法。K 均值聚类将每个目标视为在空间中有一个位置。它将目标划分为若干分区,使每个簇中的目标尽可能彼此靠近,并尽可能远离其他簇中的目标。您可以使用 imsegkmeans 函数将图像像素按值分成一个颜色空间内的若干个簇。此示例在 RGB 和 L*a*b* 颜色空间中执行图像的 k 均值聚类,以显示使用不同颜色空间如何改进分割结果。
步骤 1:读取图像
读取并显示一个用苏木精-伊红 (H&E) 染色组织的图像。这种染色方法有助于病理学家区分染成蓝-紫色和粉红色的组织类型。
he = imread("hestain.png"); imshow(he) title("H&E Image") text(size(he,2),size(he,1)+15, ... "Image courtesy of Alan Partin, Johns Hopkins University", ... FontSize=7,HorizontalAlignment="right")

步骤 2:用 K 均值聚类对 RGB 颜色空间的颜色进行分类
在 RGB 颜色空间中使用 k 均值聚类将图像分割成三个区域。对于输入图像中的每个像素,imsegkmeans 函数返回一个对应的簇标签。
将标注图像叠加显示在原始图像上。标注图像将白色、浅蓝-紫色和浅粉色区域组合在一起,这是不正确的。由于 RGB 颜色空间合并了每个通道(红、绿、蓝)内的亮度和颜色信息,因此两种不同颜色的较亮版本比这两种颜色的较暗版本更接近,也更难分割。
numColors = 3;
L = imsegkmeans(he,numColors);
B = labeloverlay(he,L);
imshow(B)
title("Labeled Image RGB")
步骤 3:将图像从 RGB 颜色空间转换为 L*a*b* 颜色空间
L*a*b* 颜色空间将图像的光度和颜色分开。这使得按颜色分割区域变得更加容易并且与亮度无关。颜色空间也更符合人类对图像中不同的白色、蓝-紫色和粉色区域的视觉感知。
L*a*b* 颜色空间是从 CIE XYZ 三色值派生的。L*a*b* 空间包含光度层 L*、色度层 a*(表示颜色落在沿红-绿轴的位置)和色度层 b*(表示颜色落在沿蓝-黄轴的位置)。所有颜色信息都在 a* 和 b* 层。
使用 rgb2lab 函数将图像转换为 L*a*b* 颜色空间。
lab_he = rgb2lab(he);
步骤 4:用 K 均值聚类对基于 a*b* 空间的颜色进行分类
要仅使用颜色信息分割图像,请将图像限制为 lab_he 中的 a* 和 b* 值。将图像转换为 single 数据类型,以便于 imsegkmeans 函数使用。使用 imsegkmeans 函数将图像像素分成三个簇。将 NumAttempts 名称-值参量的值设置为使用不同的初始簇质心位置重复聚类三次,以避免拟合局部最小值。
ab = lab_he(:,:,2:3); ab = im2single(ab); pixel_labels = imsegkmeans(ab,numColors,NumAttempts=3);
将标注图像叠加显示在原始图像上。新标注图像将白色、蓝紫色和粉色染色组织区域更清晰地区分开来。
B2 = labeloverlay(he,pixel_labels);
imshow(B2)
title("Labeled Image a*b*")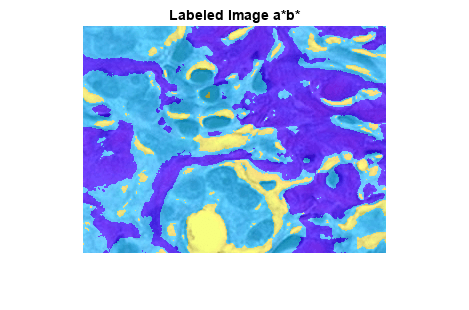
步骤 5:创建按颜色分割 H&E 图像的图像
使用 pixel_labels,您可以按颜色分离原始图像 hestain.png 中的目标,从而产生三个掩膜图像。
mask1 = pixel_labels == 1;
cluster1 = he.*uint8(mask1);
imshow(cluster1)
title("Objects in Cluster 1");
mask2 = pixel_labels == 2;
cluster2 = he.*uint8(mask2);
imshow(cluster2)
title("Objects in Cluster 2");
mask3 = pixel_labels == 3;
cluster3 = he.*uint8(mask3);
imshow(cluster3)
title("Objects in Cluster 3");
步骤 6:分割核
簇 3 中仅包含蓝色目标。请注意,有深蓝色和浅蓝色目标。您可以使用 L*a*b* 颜色空间中的 L* 层来分离深蓝色和浅蓝色。细胞核为深蓝色。
L* 层包含每个像素的亮度值。提取此簇中像素的亮度值,并使用 imbinarize 函数用全局阈值对其设置阈值。掩膜 idx_light_blue 给出了浅蓝色像素的索引。
L = lab_he(:,:,1); L_blue = L.*double(mask3); L_blue = rescale(L_blue); idx_light_blue = imbinarize(nonzeros(L_blue));
复制蓝色目标的掩膜 mask3,然后从掩膜中删除浅蓝色像素。将新掩膜应用于原始图像并显示结果。只有深蓝色细胞核可见。
blue_idx = find(mask3);
mask_dark_blue = mask3;
mask_dark_blue(blue_idx(idx_light_blue)) = 0;
blue_nuclei = he.*uint8(mask_dark_blue);
imshow(blue_nuclei)
title("Blue Nuclei")