Remove Objects from Volume Display Using 3-D Scissors
This example shows how to interactively remove unwanted regions in a 3-D display, such as a patient bed in a CT scan, by using 3-D scissors.
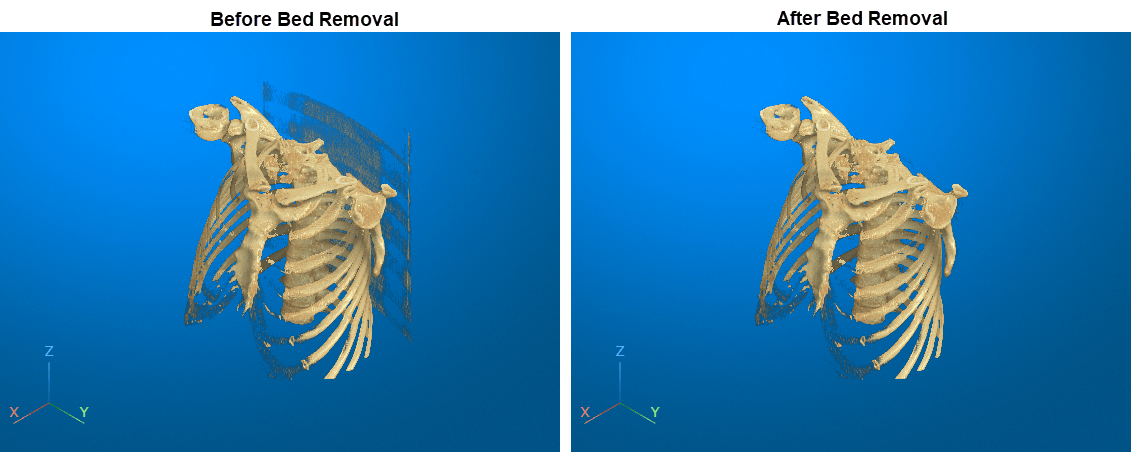
Download Image Volume Data
This example uses a subset of the Medical Segmentation Decathlon data set [1]. The subset of data includes two CT chest volumes and their corresponding label images, stored in the NIfTI file format.
Run this code to download the MedicalVolumNIfTIData.zip file from the MathWorks® website, then unzip the file. The size of the data file is approximately 76 MB.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeNIfTIData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
Specify the path to the folder that contains the downloaded and unzipped data.
dataFolder = fullfile(filepath,"MedicalVolumeNIfTIData");Specify the filenames of the volume and label images used in this example.
dataFile = fullfile(dataFolder,"lung_043.nii.gz"); labelDataFile = fullfile(dataFolder,"LabelData","lung_043.nii.gz");
Import Image Volume
Read the image data and the metadata from the image file.
V = niftiread(dataFile); info = niftiinfo(dataFile);
Extract the voxel spacing from the file metadata, and define the transformation to display the volume with correct dimensions.
voxelSize = info.PixelDimensions; sx = voxelSize(2); sy= voxelSize(1); sz = voxelSize(3); A = [sx 0 0 0; 0 sy 0 0; 0 0 sz 0; 0 0 0 1]; tform = affinetform3d(A);
Define a transparency map and colormap for this volume. The values used in this example have been determined using manual trial and error.
alpha = [0 0 0.7 0.9]; color = [0 0 0; 200 140 75; 231 208 141; 255 255 255]./255; intensity = [-3024 -700 -400 3071]; queryPoints = linspace(min(intensity),max(intensity),256); alphamap = interp1(intensity,alpha,queryPoints)'; colormap = interp1(intensity,color,queryPoints);
Display Volume
Create a viewer window in which to display the volume.
viewer = viewer3d;
Display the volume using the cinematic rendering style with the specified colormap and alphamap. To learn more about cinematic rendering, see Display Volume Using Cinematic Rendering.
vol = volshow(V, ... Parent=viewer, ... RenderingStyle="CinematicRendering", ... Transformation=tform, ... Colormap=colormap, ... Alphamap=alphamap);
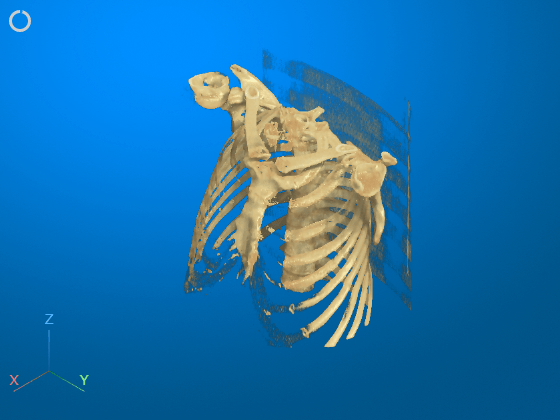
Remove Bed Using 3-D Scissors
Interactively remove the bed from the display by using the 3-D scissors tool. First, rotate the volume to the plane in which you want to define the cut region. The 3-D scissors tool removes everything within the cut out region along the direction perpendicular to the plane. For this example, click the Z axes display indicator in the bottom-left corner of the viewer to rotate the image.

To enable the 3-D scissors tool, in the viewer window toolstrip, click the scissors icon ![]() . Then, in the viewer window, draw a polygon to define a 2-D cut region. Click to place each vertex. To undo the most recent vertex placement, press Backspace. To cancel and exit the drawing operation, press Esc. Complete the drawing action by double-clicking. When you complete the drawing, the viewer automatically removes everything within the polygon.
. Then, in the viewer window, draw a polygon to define a 2-D cut region. Click to place each vertex. To undo the most recent vertex placement, press Backspace. To cancel and exit the drawing operation, press Esc. Complete the drawing action by double-clicking. When you complete the drawing, the viewer automatically removes everything within the polygon.

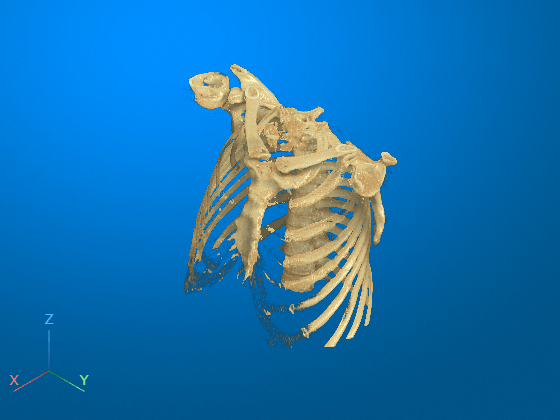
Restore the initial view position by clicking the home icon ![]() in the viewer window toolstrip. The bed has been successfully removed from the volume display. Note that you cannot undo a 3-D scissors operation. If you want to restore the full volume, reset the
in the viewer window toolstrip. The bed has been successfully removed from the volume display. Note that you cannot undo a 3-D scissors operation. If you want to restore the full volume, reset the Data property of the Volume object, or call volshow again to create a new Volume object.
References
[1] Medical Segmentation Decathlon. "Lung." Tasks. Accessed May 10, 2018. http://medicaldecathlon.com/. The Medical Segmentation Decathlon data set is provided under the CC-BY-SA 4.0 license. All warranties and representations are disclaimed. See the license for details.