导入 HDF5 文件
您可以通过多种方式从 HDF5 文件读取数据。您可以通过编程方式使用 MATLAB® 高级 HDF5 函数或低级函数的 HDF5 库命名空间。您可以通过交互方式使用导入数据实时编辑器任务或(在 MATLAB Online™ 中)导入工具。
分层数据格式版本 5 (HDF5) 是由美国国家超级计算应用中心 (NCSA) 开发的用于存储文件中科学数据的通用型机器无关标准。HDF5 广泛用于工程和科学领域,这些领域需要一种标准的数据存储方式,以便共享数据。有关 HDF5 文件格式的详细信息,请阅读 HDF Group 网站 (https://www.hdfgroup.org) 上提供的 HDF5 文档。
注意
有关导入具有单独的不兼容格式的 HDF4 文件的信息,请参阅使用高级函数导入 HDF4 文件。
使用高级 HDF5 函数导入数据
MATLAB 包含几个函数,可用于检查 HDF5 文件的内容,并将文件中的数据导入 MATLAB 工作区中。
有关如何使用这些函数的详细信息,请参阅其参考页,其中包括示例。以下各节说明一些常见的使用情形。
确定 HDF5 文件的内容
HDF5 文件可以包含数据和元数据,称为属性。HDF5 文件以类似于 UNIX® 文件系统的层次结构方式来组织数据和元数据。
在 HDF5 文件中,层次结构中的目录称为组。组可以包含其他组、数据集、属性、链接和数据类型。数据集是数据的集合,例如多维数值数组或字符串。属性是与另一实体(例如数据集)相关联的任何数据。链接类似于 UNIX 文件系统符号链接。链接是引用对象而不必制作对象副本的一种方法。
数据类型是对数据集或属性中数据的描述。数据类型说明如何解释数据集中的数据。
要快速查看 HDF5 文件的内容,请使用 h5disp 函数。
h5disp("example.h5")
HDF5 example.h5
Group '/'
Attributes:
'attr1': 97 98 99 100 101 102 103 104 105 0
'attr2': 2x2 H5T_INTEGER
Group '/g1'
Group '/g1/g1.1'
Dataset 'dset1.1.1'
Size: 10x10
MaxSize: 10x10
Datatype: H5T_STD_I32BE (int32)
ChunkSize: []
Filters: none
Attributes:
'attr1': 49 115 116 32 97 116 116 114 105 ...
'attr2': 50 110 100 32 97 116 116 114 105 ...
Dataset 'dset1.1.2'
Size: 20
MaxSize: 20
Datatype: H5T_STD_I32BE (int32)
ChunkSize: []
Filters: none
Group '/g1/g1.2'
Group '/g1/g1.2/g1.2.1'
Link: 'slink'
Type: 'soft link'
Target: 'somevalue'
Group '/g2'
Dataset 'dset2.1'
Size: 10
MaxSize: 10
Datatype: H5T_IEEE_F32BE (single)
ChunkSize: []
Filters: none
Dataset 'dset2.2'
Size: 5x3
MaxSize: 5x3
Datatype: H5T_IEEE_F32BE (single)
ChunkSize: []
Filters: none
.
.
.
要探查 HDF5 文件的层次结构组织方式,请使用 h5info 函数。h5info 函数返回包含有关 HDF5 文件的各种信息的结构体,包括文件名称。
info = h5info("example.h5")
info =
struct with fields:
Filename: 'matlabroot\matlab\toolbox\matlab\demos\example.h5'
Name: '/'
Groups: [4×1 struct]
Datasets: []
Datatypes: []
Links: []
Attributes: [2×1 struct]
通过查看 Groups 和 Attributes 字段,您可以看到该文件包含四个组和两个属性。Datasets、Datatypes 和 Links 字段都为空,表示根组不包含任何数据集、数据类型或链接。要进一步了解示例 HDF5 文件的内容,请检查 Groups 中的一个结构体。以下示例显示了此字段中第二个结构体的内容。
level2 = info.Groups(2)
level2 =
struct with fields:
Name: '/g2'
Groups: []
Datasets: [2×1 struct]
Datatypes: []
Links: []
Attributes: []
在示例文件中,名为 /g2 的组包含两个数据集。此图说明了示例 HDF5 文件组织结构的这一部分。
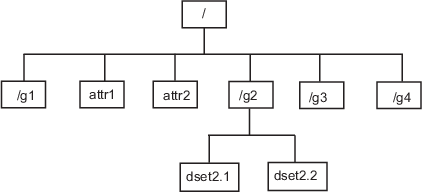
要获取有关数据集的信息(例如其名称、维度和数据类型),请查看 Datasets 字段中返回的结构体之一。
dataset1 = level2.Datasets(1)
dataset1 =
struct with fields:
Name: 'dset2.1'
Datatype: [1×1 struct]
Dataspace: [1×1 struct]
ChunkSize: []
FillValue: []
Filters: []
Attributes: []
从 HDF5 文件导入数据
要从 HDF5 文件读取数据或元数据,请使用 h5read 函数。指定 HDF5 文件的名称和数据集的名称作为参量。(要读取属性的值,必须使用 h5readatt。)
为了进行说明,此示例从 HDF5 示例文件 /g2/dset2.1 读取数据集 example.h5。
data = h5read("example.h5","/g2/dset2.1")
data =
10×1 single column vector
1.0000
1.1000
1.2000
1.3000
1.4000
1.5000
1.6000
1.7000
1.8000
1.9000
将 HDF5 数据类型映射到 MATLAB 数据类型
当 h5read 函数将数据从 HDF5 文件读入 MATLAB 工作区时,它会将 HDF5 数据类型映射到 MATLAB 数据类型,如下表所示。
| HDF5 数据类型 | h5read 输出 |
|---|---|
位字段 | 包含 8 位整数的数组 |
浮点数 | MATLAB |
整数类型,有符号和无符号 | 等效的 MATLAB 整数类型,有符号和无符号 |
不透明 |
|
引用 | 引用指向的实际数据的元胞数组,而不是引用的值 |
字符串,固定长度和可变长度 | 字符串数组 |
枚举 | 字符向量元胞数组,其中每个枚举值由对应的成员名称替换 |
复合 | 1×1 结构体数组;数据集的维度在结构体的字段中表示 |
数组 | 由数据类型与 HDF5 数组相同的值构成的数组。例如,如果数组由有符号 32 位整数构成,则 MATLAB 数组的类型为 |
MATLAB 随附的示例 HDF5 文件包含所有这些数据类型的示例。
例如,数据集 /g3/string 是字符串。
h5disp("example.h5","/g3/string")
HDF5 example.h5
Dataset 'string'
Size: 2
MaxSize: 2
Datatype: H5T_STRING
String Length: 3
Padding: H5T_STR_NULLTERM
Character Set: H5T_CSET_ASCII
Character Type: H5T_C_S1
ChunkSize: []
Filters: none
FillValue: ' '
现在从文件读取数据。MATLAB 将其以字符串数组形式返回。
s = h5read("example.h5","/g3/string")
s =
2×1 string array
"ab "
"de "
复合数据类型始终以结构体形式返回。数据集的维度在结构体的字段中表示。例如,数据集 /g3/compound2D 是复合数据类型。
h5disp("example.h5","/g3/compound2D")
HDF5 example.h5
Dataset 'compound2D'
Size: 2x3
MaxSize: 2x3
Datatype: H5T_COMPOUND
Member 'a': H5T_STD_I8LE (int8)
Member 'b': H5T_IEEE_F64LE (double)
ChunkSize: []
Filters: none
FillValue: H5T_COMPOUND
现在从文件读取数据。MATLAB 将其以结构体形式返回。
data = h5read("example.h5","/g3/compound2D")
data =
struct with fields:
a: [2×3 int8]
b: [2×3 double]
使用低级 HDF5 函数导入数据
MATLAB 支持通过低级函数直接访问 HDF5 库中的数十个函数,这些低级函数对应于 HDF5 库中的函数。通过这种方式,您就可以从 MATLAB 访问 HDF5 库的功能,例如使用 HDF5 子集功能和编写复数数据类型。有关详细信息,请参阅Export Data Using MATLAB Low-Level HDF5 Functions。
以交互方式从 HDF5 文件读取数据
此示例说明如何使用导入数据任务来浏览 HDF5 文件的结构,从文件中导入数据,然后分析和可视化数据。
浏览和导入数据
此示例附带的 HDF5 文件包含来自 NASA Precipitation Processing System (PPS) 的一系列降水概率估计值。
通过选择实时编辑器选项卡上的任务 > 导入数据,在实时编辑器中打开导入数据任务。在文件字段中输入 HDF5 文件的名称 nasa_pps.h5。使用任务浏览数据的结构,包括组、数据集和属性。该文件包含组 S1,其中包含三个数据集。probabilityOfPrecip 数据集包含给定位置的降水概率估计值;Latitude 和 Longitude 数据集包含这些概率估计值的位置。probabilityOfPrecip 数据集的属性包括 units,其值为 'percent'。
选择并导入三个数据集的数据和属性。
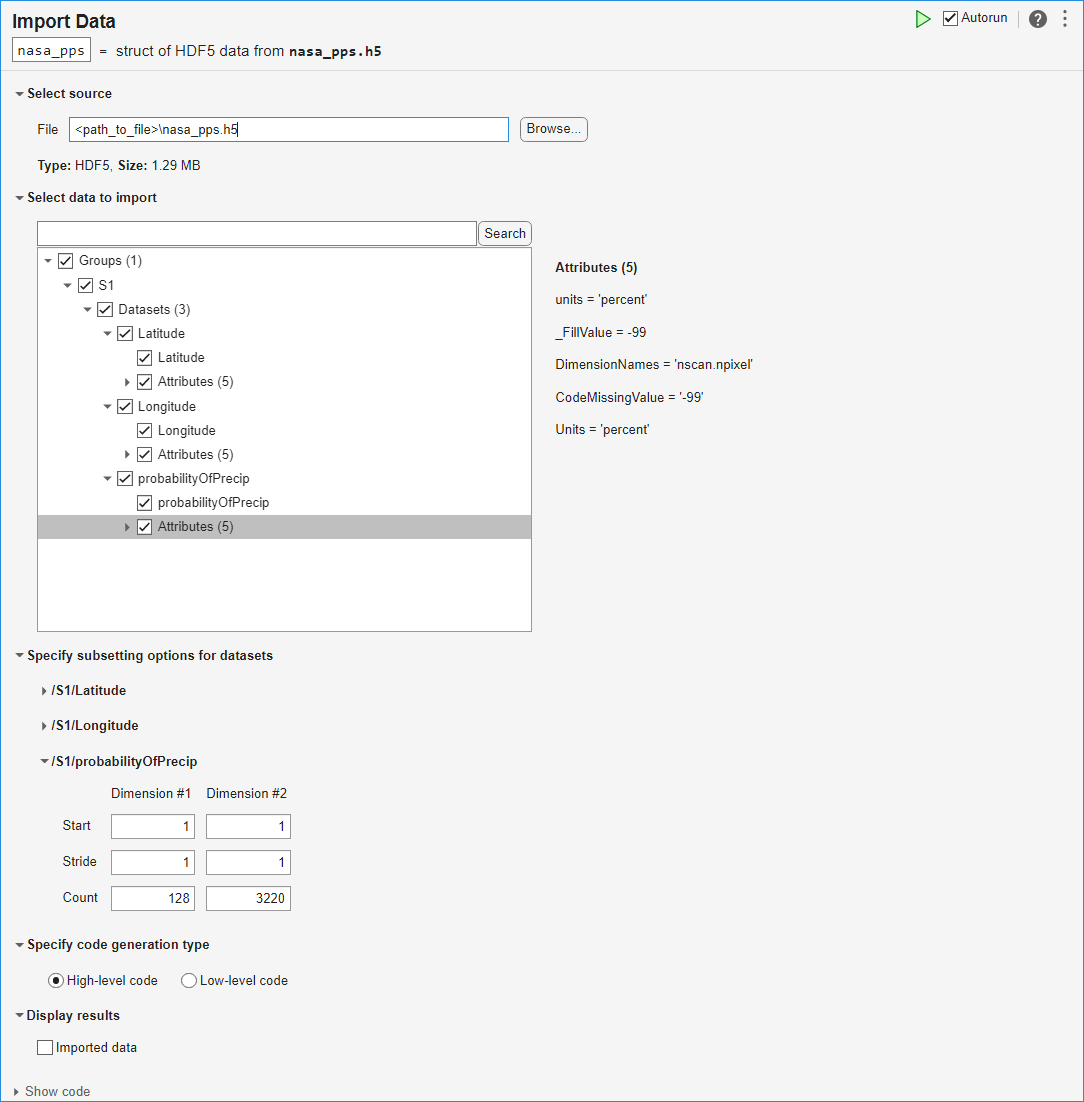
要查看此任务生成的代码,请点击任务参数区域底部的显示代码展开任务显示。
% Create a structure to store imported HDF5 data nasa_pps = struct(); filename = "nasa_pps.h5"; nasa_pps.Groups(1).Name = "S1"; nasa_pps.Groups(1).Datasets(1).Name = "Latitude"; nasa_pps.Groups(1).Datasets(1).Value = h5read(filename, "/S1/Latitude"); nasa_pps.Groups(1).Datasets(2).Name = "Longitude"; nasa_pps.Groups(1).Datasets(2).Value = h5read(filename, "/S1/Longitude"); nasa_pps.Groups(1).Datasets(3).Name = "probabilityOfPrecip"; nasa_pps.Groups(1).Datasets(3).Value = h5read(filename, "/S1/probabilityOfPrecip"); nasa_pps.Groups(1).Datasets(1).Attributes(1).Name = "units"; nasa_pps.Groups(1).Datasets(1).Attributes(1).Value = h5readatt(filename, "/S1/Latitude", "units"); nasa_pps.Groups(1).Datasets(1).Attributes(2).Name = "DimensionNames"; nasa_pps.Groups(1).Datasets(1).Attributes(2).Value = h5readatt(filename, "/S1/Latitude", "DimensionNames"); nasa_pps.Groups(1).Datasets(1).Attributes(3).Name = "Units"; nasa_pps.Groups(1).Datasets(1).Attributes(3).Value = h5readatt(filename, "/S1/Latitude", "Units"); nasa_pps.Groups(1).Datasets(1).Attributes(4).Name = "_FillValue"; nasa_pps.Groups(1).Datasets(1).Attributes(4).Value = h5readatt(filename, "/S1/Latitude", "_FillValue"); nasa_pps.Groups(1).Datasets(1).Attributes(5).Name = "CodeMissingValue"; nasa_pps.Groups(1).Datasets(1).Attributes(5).Value = h5readatt(filename, "/S1/Latitude", "CodeMissingValue"); nasa_pps.Groups(1).Datasets(2).Attributes(1).Name = "units"; nasa_pps.Groups(1).Datasets(2).Attributes(1).Value = h5readatt(filename, "/S1/Longitude", "units"); nasa_pps.Groups(1).Datasets(2).Attributes(2).Name = "DimensionNames"; nasa_pps.Groups(1).Datasets(2).Attributes(2).Value = h5readatt(filename, "/S1/Longitude", "DimensionNames"); nasa_pps.Groups(1).Datasets(2).Attributes(3).Name = "Units"; nasa_pps.Groups(1).Datasets(2).Attributes(3).Value = h5readatt(filename, "/S1/Longitude", "Units"); nasa_pps.Groups(1).Datasets(2).Attributes(4).Name = "_FillValue"; nasa_pps.Groups(1).Datasets(2).Attributes(4).Value = h5readatt(filename, "/S1/Longitude", "_FillValue"); nasa_pps.Groups(1).Datasets(2).Attributes(5).Name = "CodeMissingValue"; nasa_pps.Groups(1).Datasets(2).Attributes(5).Value = h5readatt(filename, "/S1/Longitude", "CodeMissingValue"); nasa_pps.Groups(1).Datasets(3).Attributes(1).Name = "units"; nasa_pps.Groups(1).Datasets(3).Attributes(1).Value = h5readatt(filename, "/S1/probabilityOfPrecip", "units"); nasa_pps.Groups(1).Datasets(3).Attributes(2).Name = "_FillValue"; nasa_pps.Groups(1).Datasets(3).Attributes(2).Value = h5readatt(filename, "/S1/probabilityOfPrecip", "_FillValue"); nasa_pps.Groups(1).Datasets(3).Attributes(3).Name = "DimensionNames"; nasa_pps.Groups(1).Datasets(3).Attributes(3).Value = h5readatt(filename, "/S1/probabilityOfPrecip", "DimensionNames"); nasa_pps.Groups(1).Datasets(3).Attributes(4).Name = "CodeMissingValue"; nasa_pps.Groups(1).Datasets(3).Attributes(4).Value = h5readatt(filename, "/S1/probabilityOfPrecip", "CodeMissingValue"); nasa_pps.Groups(1).Datasets(3).Attributes(5).Name = "Units"; nasa_pps.Groups(1).Datasets(3).Attributes(5).Value = h5readatt(filename, "/S1/probabilityOfPrecip", "Units"); clear filename
组织和准备数据
使用冒号运算符 (:) 将三个数据集的数据提取为列向量。
lats = nasa_pps.Groups.Datasets(1).Value(:); lons = nasa_pps.Groups.Datasets(2).Value(:); probs = nasa_pps.Groups.Datasets(3).Value(:);
清理数据以备绘图。这三个数据集中的有效索引符合以下条件:
probs中的值在区间 [0, 100] 内。lats中的值在区间 [–90, 90] 内。lons中的值在区间 [–180, 180] 内。
通过使用逻辑索引,仅保留三个数据集中有效索引处的值。此步骤删除无效的经度和纬度值以及概率估计值的无效百分比值。
isValid = 0<=probs & probs<=100 & abs(lats)<=90 & abs(lons)<=180; probs = probs(isValid); lats = lats(isValid); lons = lons(isValid);
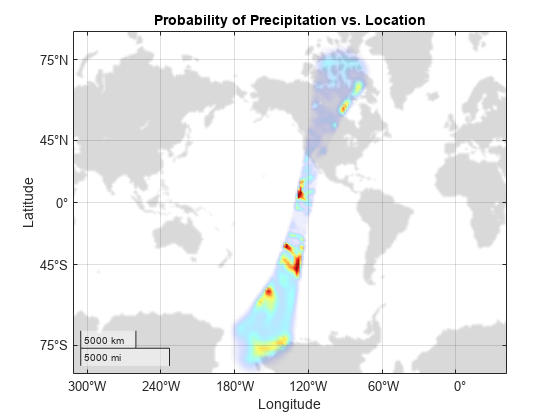
对数据绘图
为数据集中的有效数据创建一个地理密度图。“较暖”的颜色表示较高的降水概率。
g = geodensityplot(lats,lons,probs,FaceColor="interp"); geobasemap grayland geolimits([-80 80],[-180 -90]) title("Probability of Precipitation vs. Location") colormap turbo
感谢
全球降水数据由 NASA Precipitation Processing System 提供,来自其网站 https://gpm.nasa.gov/data/directory。
使用动态加载的过滤器读取 HDF5 数据集
MATLAB 支持使用动态加载的过滤器读写 HDF5 数据集。HDF Group 在其网站的过滤器部分维护已注册过滤器的列表。
要读取使用用户定义的第三方过滤器写入的数据集,请按照以下步骤操作:
在您的系统上将 HDF5 过滤器插件安装为共享库或 DLL。
设置
HDF5_PLUGIN_PATH环境变量以指向插件的本地安装,然后启动 MATLAB:在 Windows® 上 - 使用系统属性 > 高级 > 环境变量设置环境变量,然后启动 MATLAB。
在 Linux® 和 Mac 上 - 从终端设置环境变量,然后从同一终端启动 MATLAB。
在完成这些步骤后,您可以使用高级或低级 MATLAB HDF5 函数来读取和访问已使用第三方过滤器压缩的数据集。有关详细信息,请参阅 HDF Group 网站上的 HDF5 动态加载的过滤器。
仅限 Linux 用户:使用 MATLAB HDF5 共享库重新编译过滤器插件
从 R2021b 开始,在某些情况下,使用需要回调核心 HDF5 库函数的过滤器插件的 Linux 用户必须使用附带的 MATLAB HDF5 共享库 /matlab/bin/glnxa64/libhdf5.so.x.x.x 重新编译插件。如果您不使用此版本的共享库重新编译插件,则可能会遇到各种问题,包括未定义的行为,甚至可能导致系统崩溃。有关详细信息,请参阅使用 MATLAB HDF5 共享库或 GNU 导出映射在 Linux 上编译 HDF5 过滤器插件。