Read, Process, and View Ultrasound Data
This example shows how to import and display a 2-D multiframe ultrasound series, and how to apply a denoising filter to each frame of the series.
You can use the medicalImage object to import image data and metadata from 2-D medical images and series of images related by time. In this example, you use the properties and the extractFrame object function of a medicalImage object to work with a multiframe echocardiogram ultrasound series.
Read Ultrasound Image Series
Specify the name of an echocardiogram ultrasound series contained in a DICOM file.
filename = "heartUltrasoundSequenceVideo.dcm";Read the metadata and image data from the file by creating a medicalImage object. The image data is stored in the Pixels property. Each frame of the image series is a 600-by-800-by-3 pixel RGB image. The FrameTime property indicates that each frame has a duration of 33.333 milliseconds. The NumFrames property indicates that the series has a total of 116 image frames.
medImg = medicalImage(filename)
medImg =
medicalImage with properties:
Pixels: [600×800×116×3 uint8]
Colormap: []
SpatialUnits: "unknown"
FrameTime: 33.3330
NumFrames: 116
PixelSpacing: [1 1]
Modality: 'US'
WindowCenter: []
WindowWidth: []
Display Ultrasound Image Frames as Montage
Display the frames in the Pixels property of medImg as a montage.
montage(medImg)

Display Ultrasound Series as Video
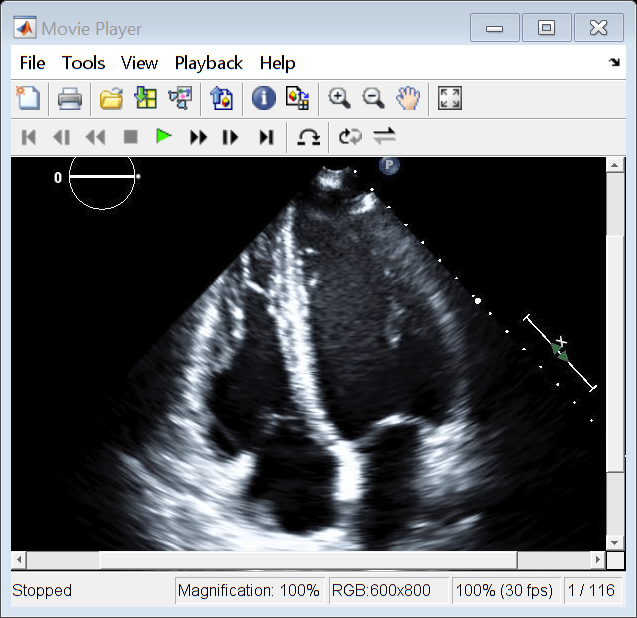
Open the Video Viewer app to view the ultrasound series as a video by using the implay function. The implay function automatically sets the frame rate using the FrameTime property of medImg.
implay(medImg)
Reduce Speckle Noise
Reduce the noise in the ultrasound image series by applying a speckle-reducing anisotropic diffusion filter to each frame. Extract each frame from the medicalImage object by using the extractFrame object function, and convert the frame from RGB to grayscale. Apply the filter by using the specklefilt function. Specify the DegreeOfSmoothing and NumIterations name-value arguments to control the smoothing parameters.
[h,w,d,c] = size(medImg.Pixels); pixelsSmoothed = zeros(h,w,d); for i = 1:d Ii = extractFrame(medImg,i); Ii = im2double(im2gray(Ii)); pixelsSmoothed(:,:,i) = specklefilt(Ii,DegreeOfSmoothing=0.6,NumIterations=50); end
Store Smoothed Data as Medical Image Object
Create a new medicalImage object that contains the smoothed image pixels. Use the property values from the original file to maintain the correct frame information.
medImgSmoothed = medImg; medImgSmoothed.Pixels = pixelsSmoothed
medImgSmoothed =
medicalImage with properties:
Pixels: [600×800×116 double]
Colormap: []
SpatialUnits: "unknown"
FrameTime: 33.3330
NumFrames: 116
PixelSpacing: [1 1]
Modality: 'US'
WindowCenter: []
WindowWidth: []
View the first frame of the smoothed image series by using the montage function with the Indices name-value argument set to 1.
figure montage(medImgSmoothed,Indices=1)

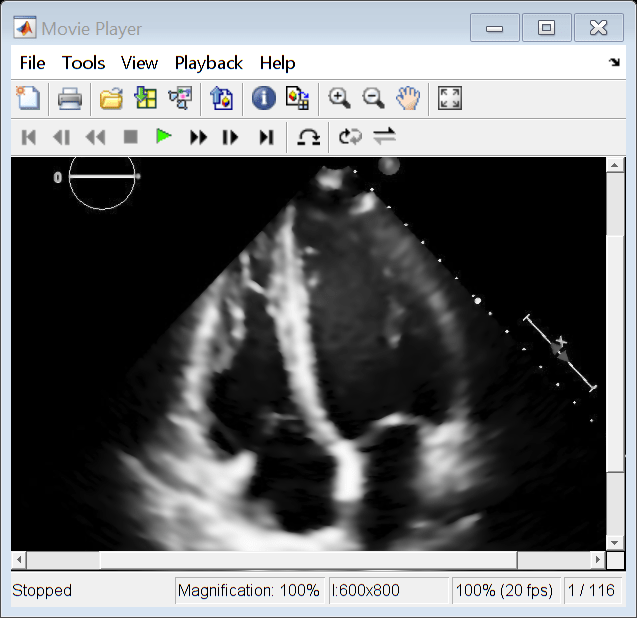
Display the smoothed image data as a video.
implay(pixelsSmoothed)
See Also
medicalImage | extractFrame | specklefilt | montage | Video Viewer