compare
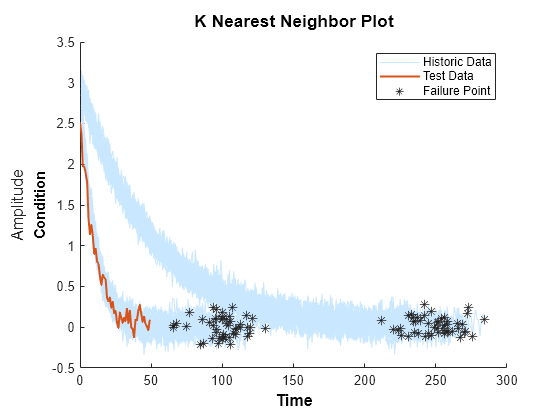
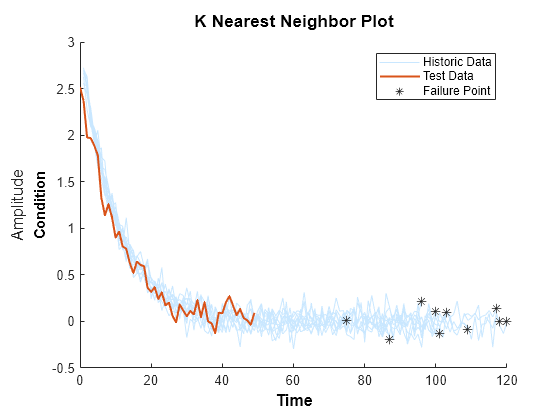
Compare test data to historical data ensemble for similarity models
Description
compare(___,
specifies plotting options using one or more name-value pair arguments.Name,Value)
Examples
Input Arguments
Name-Value Arguments
Version History
Introduced in R2018a