distanceProfile
Compute distance profile between query subsequence and all other subsequences of a single-variable or multivariable time series
Since R2024b
Syntax
Description
Return Distance Profile
DP = distanceProfile(X,len,loc)X and every
subsequence in X with the same length len.
If
Xis a vector, then the software treats it as a single channelIf
Xis a matrix, then you can control whether the software computes the distance profile for each time series channel individually or cumulatively using theTypename-value argument.
The query begins at the time series position loc. The query
subsequence is therefore defined by whether X is a vector or a
matrix:
Vector —
X(loc:loc+len-1)Matrix with K columns —
X(loc:loc+len-1,k).
[___] = distanceProfile(___,Name=Value)
specifies options using one or more name-value arguments in addition to the arguments in
previous syntaxes. For example, to exclude matches near the query starting position, set
ExcludeTrivialMatches to true.
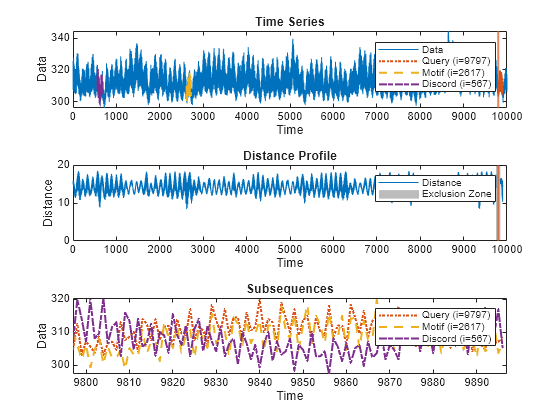
Plot Distance Profile
distanceProfile(___) plots an interactive plot of the
distance profile, with overlays for the query, the motif (best match to query), and the
discord (worst match to query). You can move the vertical selection lines in the plot to
find the top motif and discord of any other data segments in the time series.
You can use this syntax with any of the previous input-argument combinations.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Yeh, Chin-Chia Michael, et al. “Matrix Profile I: All Pairs Similarity Joins for Time Series: A Unifying View That Includes Motifs, Discords and Shapelets.” 2016 IEEE 16th International Conference on Data Mining (ICDM), IEEE, 2016, pp. 1317–22. DOI.org (Crossref), https://doi.org/10.1109/ICDM.2016.0179.
Extended Capabilities
Version History
Introduced in R2024b