addkineticlaw (reaction)
Create kinetic law object and add to reaction object
Syntax
kineticlawObj =
addkineticlaw(reactionObj, 'KineticLawNameValue')
kineticlawObj=
addkineticlaw(..., 'PropertyName', PropertyValue,
...)
Arguments
reactionObj | Reaction object. Enter a variable name for a reaction object. |
KineticLawNameValue | Property to select the type of kinetic law object to create.
For built-in kinetic law, valid values are:
Find valid
|
Description
kineticlawObj =
addkineticlaw(reactionObj, 'KineticLawNameValue')KineticLaw object to
the reactionObj.
In the kinetic law object, this method assigns a name (KineticLawNameValueKineticLawName and assigns the
reaction object to the property Parent. In the
reaction object, this method assigns the kinetic law object to the
property KineticLaw.
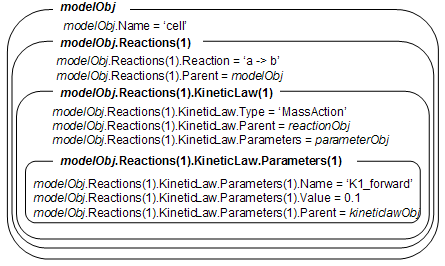
modelObj = sbiomodel('cell');
reactionObj = addreaction(modelObj, 'a -> b');
kineticlawObj = addkineticlaw(reactionObj, 'MassAction');
parameterObj = addparameter(kineticlawObj, 'K1_forward', 0.1);
set(kineticlawObj, ParameterVariableName, 'K1_forward');
KineticLawNameValue
constructs a kinetic law object, kineticlawObj=
addkineticlaw(..., 'PropertyName', PropertyValue,
...)kineticlawObjkineticlawObjset.
The kineticlawObj properties are listed in Property Summary.
Note
To define a Hill kinetic rate equation with a non-integer exponent that is compatible with DimensionalAnalysis, see Define a Custom Hill Kinetic Law that Works with Dimensional Analysis.
Property Summary
Properties for kinetic law objects
| Notes | HTML text describing SimBiology object |
| Type | Display SimBiology object type |
| UserData | Specify data to associate with object |
Examples
See Also
Version History
Introduced in R2006a