SimBiology.Variant
Store alternate component values
Description
The SimBiology.Variant object stores the names and values of model
components and allows you to use the values stored in a variant object as the alternate value
to be applied during a simulation. You can store values for species InitialAmount, parameter Value, and compartment Capacity in a variant object. Simulating using a variant does not alter the
model component values. The values specified in the variant temporarily apply during
simulation.
Using one or more variant objects associated with a model allows you to evaluate model
behavior during simulation, with different values for the various model components without
having to search and replace these values, or having to create additional models with these
values. If you determine that the values in a variant object accurately define your model, you
can permanently replace the values in your model with the values stored in the variant object,
using the commit function.
To use a variant in a simulation you must add the variant object to the model object and
set the Active property of the variant to true. Set the
Active property to true if you always want the variant to be applied
before simulating the model. You can also enter the variant object as an argument to sbiosimulate; this applies the variant only for the current simulation and
supersedes any active variant objects on the model.
When there are multiple active variant objects on a model, if there are duplicate
specifications for a property's value, the last occurrence for the property value in the array
of variants, is used during simulation. You can find out which variant is applied last by
looking at the indices of the variant objects stored on the model. Similarly, in the
Content property, if there are duplicate specifications for a property's
value, the last occurrence for the property in the Content property, is
used during simulation.
Use dot notation to query the object properties or change properties that are not
read-only. You can also use the get and set commands.
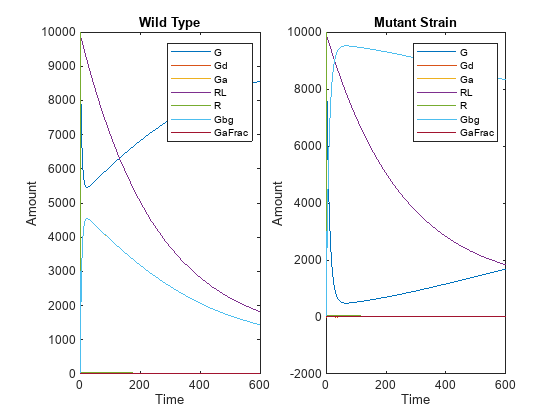
The SimBiology Model Builder app also enables you to add variants to your model and edit them. For an example, see Represent Biological Variability Using Variants.
Creation
Use sbiovariant to create a
SimBiology.Variant object. Use addvariant (model) to add a SimBiology.Variant object to a
SimBiology model.
Properties
Object Functions
addcontent (variant) | Append content to variant object |
commit (variant) | Commit variant contents to model |
copyobj | Copy SimBiology object and its children |
delete | Delete SimBiology object |
display | Display summary of SimBiology object |
get | Get SimBiology object properties |
rename | Rename SimBiology model component and update expressions |
rmcontent (variant) | Remove contents from variant object |
set | Set SimBiology object properties |
verify (model, variant) | Validate and verify SimBiology model |