predict
Description
yFit = predict(Mdl,X,Name,Value)'IncludeInteractions',true specifies to include interaction terms in
computations.
[
also returns the standard deviations and prediction intervals of the response variable,
evaluated at each observation in the predictor data yFit,ySD,yInt] = predict(___)X, using any of the
input argument combinations in the previous syntaxes. This syntax is valid only when you
specify 'FitStandardDeviation' of fitrgam as
true for training Mdl and the IsStandardDeviationFit property of Mdl is
true.
Examples
Train a generalized additive model using training samples, and then predict the test sample responses.
Load the patients data set.
load patientsCreate a table that contains the predictor variables (Age, Diastolic, Smoker, Weight, Gender, SelfAssessedHealthStatus) and the response variable (Systolic).
tbl = table(Age,Diastolic,Smoker,Weight,Gender,SelfAssessedHealthStatus,Systolic);
Randomly partition observations into a training set and a test set. Specify a 10% holdout sample for testing.
rng('default') % For reproducibility cv = cvpartition(size(tbl,1),'HoldOut',0.10);
Extract the training and test indices.
trainInds = training(cv); testInds = test(cv);
Train a univariate GAM that contains the linear terms for the predictors in tbl.
Mdl = fitrgam(tbl(trainInds,:),'Systolic')Mdl =
RegressionGAM
PredictorNames: {'Age' 'Diastolic' 'Smoker' 'Weight' 'Gender' 'SelfAssessedHealthStatus'}
ResponseName: 'Systolic'
CategoricalPredictors: [3 5 6]
ResponseTransform: 'none'
Intercept: 122.7444
IsStandardDeviationFit: 0
NumObservations: 90
Properties, Methods
Mdl is a RegressionGAM model object.
Predict responses for the test set.
yFit = predict(Mdl,tbl(testInds,:));
Create a table containing the observed response values and the predicted response values.
table(tbl.Systolic(testInds),yFit, ... 'VariableNames',{'Observed Value','Predicted Value'})
ans=10×2 table
Observed Value Predicted Value
______________ _______________
124 126.58
121 123.95
130 116.72
115 117.35
121 117.45
116 118.5
123 126.16
132 124.14
125 127.36
124 115.99
Predict responses for new observations using a generalized additive model that contains both linear and interaction terms for predictors. Use a memory-efficient model object, and specify whether to include interaction terms when predicting responses.
Load the carbig data set, which contains measurements of cars made in the 1970s and early 1980s.
load carbigSpecify Acceleration, Displacement, Horsepower, and Weight as the predictor variables (X) and MPG as the response variable (Y).
X = [Acceleration,Displacement,Horsepower,Weight]; Y = MPG;
Partition the data set into two sets: one containing training data, and the other containing new, unobserved test data. Reserve 10 observations for the new test data set.
rng('default')
n = size(X,1);
newInds = randsample(n,10);
inds = ~ismember(1:n,newInds);
XNew = X(newInds,:);
YNew = Y(newInds);Train a GAM that contains all the available linear and interaction terms in X.
Mdl = fitrgam(X(inds,:),Y(inds),'Interactions','all');
Mdl is a RegressionGAM model object.
Conserve memory by reducing the size of the trained model.
CMdl = compact(Mdl); whos('Mdl','CMdl')
Name Size Bytes Class Attributes CMdl 1x1 1255766 classreg.learning.regr.CompactRegressionGAM Mdl 1x1 1289882 RegressionGAM
CMdl is a CompactRegressionGAM model object.
Predict the responses using both linear and interaction terms, and then using only linear terms. To exclude interaction terms, specify 'IncludeInteractions',false.
yFit = predict(CMdl,XNew);
yFit_nointeraction = predict(CMdl,XNew,'IncludeInteractions',false);Create a table containing the observed response values and the predicted response values.
t = table(YNew,yFit,yFit_nointeraction, ... 'VariableNames',{'Observed Response', ... 'Predicted Response','Predicted Response Without Interactions'})
t=10×3 table
Observed Response Predicted Response Predicted Response Without Interactions
_________________ __________________ _______________________________________
27.9 23.04 23.649
NaN 37.163 35.779
NaN 25.876 21.978
13 12.786 14.141
36 28.889 27.281
19.9 22.199 18.451
24.2 23.995 24.885
12 14.247 13.982
38 33.797 33.528
13 12.225 11.127
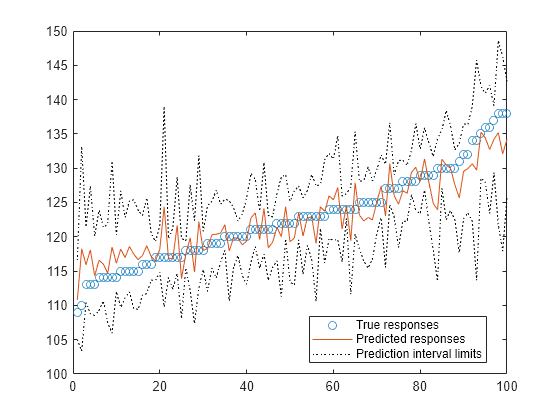
Train a generalized additive model (GAM), and then compute and plot the prediction intervals of response values.
Load the patients data set.
load patientsCreate a table that contains the predictor variables (Age, Diastolic, Smoker, Weight, Gender, SelfAssessedHealthStatus) and the response variable (Systolic).
tbl = table(Age,Diastolic,Smoker,Weight,Gender,SelfAssessedHealthStatus,Systolic);
Train a univariate GAM that contains the linear terms for the predictors in tbl. Specify the FitStandardDeviation name-value argument as true so that you can use the trained model to compute prediction intervals. A recommended practice is to use optimal hyperparameters when you fit the standard deviation model for the accuracy of the standard deviation estimates. Specify 'OptimizeHyperparameters' as 'all-univariate'. For reproducibility, use the 'expected-improvement-plus' acquisition function. Specify 'ShowPlots' as false and 'Verbose' as 0 to disable plot and message displays, respectively.
rng('default') % For reproducibility Mdl = fitrgam(tbl,'Systolic','FitStandardDeviation',true, ... 'OptimizeHyperparameters','all-univariate', ... 'HyperparameterOptimizationOptions',struct('AcquisitionFunctionName','expected-improvement-plus', ... 'ShowPlots',false,'Verbose',0))
Mdl =
RegressionGAM
PredictorNames: {'Age' 'Diastolic' 'Smoker' 'Weight' 'Gender' 'SelfAssessedHealthStatus'}
ResponseName: 'Systolic'
CategoricalPredictors: [3 5 6]
ResponseTransform: 'none'
Intercept: 122.7800
IsStandardDeviationFit: 1
NumObservations: 100
HyperparameterOptimizationResults: [1×1 BayesianOptimization]
Properties, Methods
Mdl is a RegressionGAM model object that uses the best estimated feasible point. The best estimated feasible point indicates the set of hyperparameters that minimizes the upper confidence bound of the objective function value based on the underlying objective function model of the Bayesian optimization process. For more details on the optimization process, see Optimize GAM Using OptimizeHyperparameters.
Predict responses for the training data in tbl, and compute the 99% prediction intervals of the response variable. Specify the significance level ('Alpha') as 0.01 to set the confidence level of the prediction intervals to 99%.
[yFit,~,yInt] = predict(Mdl,tbl,'Alpha',0.01);Plot the sorted true responses together with the predicted responses and prediction intervals.
figure yTrue = tbl.Systolic; [sortedYTrue,I] = sort(yTrue); plot(sortedYTrue,'o') hold on plot(yFit(I)) plot(yInt(I,1),'k:') plot(yInt(I,2),'k:') legend('True responses','Predicted responses', ... 'Prediction interval limits','Location','best') hold off
Input Arguments
Generalized additive model, specified as a RegressionGAM
or a CompactRegressionGAM model object.
Predictor data, specified as a numeric matrix or table.
Each row of X corresponds to one observation, and each column corresponds to one variable.
For a numeric matrix:
The variables that make up the columns of
Xmust have the same order as the predictor variables that trainedMdl.If you trained
Mdlusing a table, thenXcan be a numeric matrix if the table contains all numeric predictor variables.
For a table:
If you trained
Mdlusing a table (for example,Tbl), then all predictor variables inXmust have the same variable names and data types as those inTbl. However, the column order ofXdoes not need to correspond to the column order ofTbl.If you trained
Mdlusing a numeric matrix, then the predictor names inMdl.PredictorNamesand the corresponding predictor variable names inXmust be the same. To specify predictor names during training, use the'PredictorNames'name-value argument. All predictor variables inXmust be numeric vectors.Xcan contain additional variables (response variables, observation weights, and so on), butpredictignores them.predictdoes not support multicolumn variables or cell arrays other than cell arrays of character vectors.
Data Types: table | double | single
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'Alpha',0.01,'IncludeInteractions',false specifies the
confidence level as 99% and excludes interaction terms from computations.
Significance level for the confidence level of the prediction intervals
yInt, specified as a numeric scalar in the range
[0,1]. The confidence level of yInt is equal
to 100(1 – Alpha)%.
This argument is valid only when the IsStandardDeviationFit property of Mdl is
true. Specify the 'FitStandardDeviation' name-value argument of
fitrgam as true to fit the model for the
standard deviation.
Example: 'Alpha',0.01 specifies to return 99% prediction
intervals.
Data Types: single | double
Flag to include interaction terms of the model, specified as true or
false.
The default 'IncludeInteractions' value is true if Mdl contains interaction terms. The value must be false if the model does not contain interaction terms.
Example: 'IncludeInteractions',false
Data Types: logical
Output Arguments
Predicted responses, returned as a column vector of length n,
where n is the number of observations in the predictor data
X.
Standard deviations of the response variable, evaluated at each observation in the
predictor data X, returned as a column vector of length
n, where n is the number of observations in
X. The ith element ySD(i)
contains the standard deviation of the ith response for the
ith observation X(i,:)Mdl.
This argument is valid only when the IsStandardDeviationFit property of Mdl is
true. Specify the 'FitStandardDeviation' name-value argument of fitrgam
as true to fit the model for the standard deviation.
Prediction intervals of the response variable, evaluated at each observation in the
predictor data X, returned as an n-by-2 matrix,
where n is the number of observations in X. The
ith row yInt(i,:) contains the
100(1– prediction interval of the
Alpha)%ith response for the ith observation
X(i,:)Alpha value
is the probability that the prediction interval does not contain the true response value
for X(i,:)yInt contains the lower limits of the prediction intervals, and
the second column contains the upper limits.
This argument is valid only when the IsStandardDeviationFit property of Mdl is
true. Specify the 'FitStandardDeviation' name-value argument of fitrgam
as true to fit the model for the standard deviation.
Algorithms
predict returns the predicted responses (yFit)
and, optionally, the standard deviations (ySD) and prediction intervals
(yInt) of the response variable, estimated at each observation in
X.
A Generalized Additive Model (GAM) for Regression
assumes that the response variable y follows the normal distribution with
mean μ and standard deviation σ. If you specify
'FitStandardDeviation' of fitrgam as
false (default), then fitrgam trains a model for
μ. If you specify 'FitStandardDeviation' as
true, then fitrgam trains an additional model for
σ and sets the IsStandardDeviationFit property of
the GAM object to true. The outputs yFit and
ySD correspond to the estimated mean μ and
standard deviation σ, respectively.
Version History
Introduced in R2021a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)