cmddenoise
Interval-dependent denoising
Syntax
Description
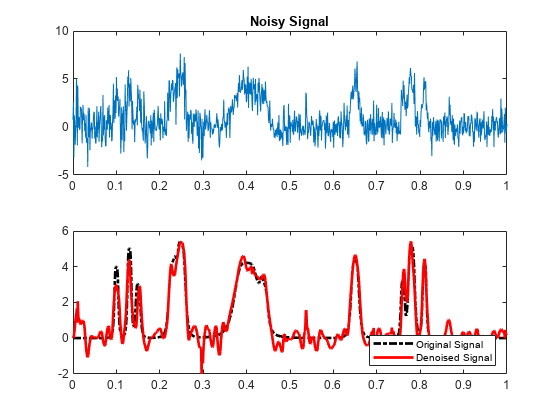
sigden = cmddenoise(sig,wname,level)sigden, obtained from an
interval-dependent denoising of the signal, sig, using the
orthogonal or biorthogonal wavelet and scaling filters,
wname. cmddenoise thresholds the
wavelet (detail) coefficients down to level, level, and
reconstructs a signal approximation using the modified detail coefficients.
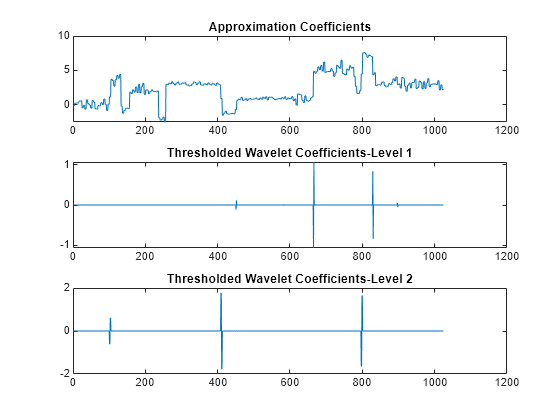
cmddenoise partitions the signal into intervals based
on variance change points in the first level detail coefficients and thresholds
each interval separately. The location and number of variance change points are
automatically selected using a penalized contrast function [2]. The minimum delay between change points is 10 samples. Thresholds are
obtained using a minimax threshold rule and soft thresholding is used to modify
the wavelet coefficients [1].
sigden = cmddenoise(sig,wname,level,sorh,nb_inter,thrParamsIn)sigden, with the denoising
intervals and corresponding thresholds specified as a cell array of
matrices with length equal to level. Each element
of the cell array contains the interval and threshold information
for the corresponding level of the wavelet transform. The elements
of thrParamsIn are N-by-3 matrices with N equal
to the number of intervals. The 1st and 2nd columns contain the beginning
and ending indices of the intervals and the 3rd column contains the
corresponding threshold value. If you specify thrParamsIn, cmddenoise ignores
the value of nb_inter.
[ returns a cell array, sigden,coefs,thrParamsOut]
= cmddenoise(___)thrParamsOut,
with length equal to level. Each element of thrParamsOut is
an N-by-3 matrix. The row dimension of the matrix elements is the
number of intervals and is determined by the value of the input arguments.
Each row of the matrix contains the beginning and end points (indices)
of the thresholded interval and the corresponding threshold value.
[ returns
a cell array, sigden,coefs,thrParamsOut,int_DepThr_Cell]
= cmddenoise(sig,wname,level,sorh,nb_inter)int_DepThr_Cell, with length equal
to 6. int_DepThr_Cell contains interval and threshold
information assuming the number of change points ranges from 0 to
5. The N-th element of int_DepThr_Cell is a N-by-3
matrix containing the interval information assuming N-1 change points.
Each row of the matrix contains the beginning and end points (indices)
of the thresholded interval and the corresponding threshold value.
Attempting to output int_DepThr_Cell if you use
the input argument, thrParamsIn, results in an
error.
[ returns
the optimal number of signal intervals based on the estimated variance
change points in the level-1 detail coefficients. To estimate the
number of change points, sigden,coefs,thrParamsOut,int_DepThr_Cell,BestNbofInt]
= cmddenoise(sig,wname,level,sorh,nb_inter)cmddenoise assumes the
total number is less than or equal to 6 and uses a penalized contrast [2].
Attempting to output BestNbofInt if you use the
input argument, thrParamsIn, results in an error.
Examples
Input Arguments
Output Arguments
References
[1] Donoho, D. and Johnstone, I. “Ideal spatial adaptation by wavelet shrinkage”, Biometrika, 1994, 81,3, 425–455.
[2] Lavielle, M. “Detection of multiple changes in a sequence of dependent variables”, Stochastic Processes and their Applications, 1999, 83, 79–102.
Version History
Introduced in R2010a