eqn =
搜索
Three former MathWorks employees, Steve Wilcockson, David Bergstein, and Gareth Thomas, joined the ArrayCast pod cast to discuss their work on array based languages. At the end of the episode, Steve says,
> It's a little known fact about MATLAB. There's this thing, Gareth has talked about the community. One of the things MATLAB did very, very early was built the MATLAB community, the so-called MATLAB File Exchange, which came about in the early 2000s. And it was where people would share code sets, M files, et cetera. This was long before GitHub came around. This was well ahead of its time. And I think there are other places too, where MATLAB has delivered cultural benefits over and above the kind of core programming and mathematical capabilities too. So, you know, MATLAB Central, File Exchange, very much saw the future.
Listen here: The ArrayCast, Episode 79, May 10, 2024.
This topic is for discussing highlights to the current R2025a Pre-release.
So you've downloaded the R2025a pre-release, tried Dark mode and are wondering what else is new. A lot! A lot is new!
One thing I am particularly happy about is the fact that Apple Accelerate is now the default BLAS on Apple Silicon machines. Check it out by doing
>> version -blas
ans =
'Apple Accelerate BLAS (ILP64)'
If you compare this to R2024b that is using OpenBLAS you'll see some dramatic speed-ups in some areas. For example, I saw up to 3.7x speed-up for matrix-matrix multiplication on my M2 Mabook Pro and 2x faster LU factorisation.
Details regarding my experiments are in this blog post Life in the fast lane: Making MATLAB even faster on Apple Silicon with Apple Accelerate » The MATLAB Blog - MATLAB & Simulink . Back then you had to to some trickery to switch to Apple Accelerate, now its the default.
I just published a blog post called "The Story of TIMEIT." I've been thinking about writing something like this ever since Mike Croucher's tic/toc blog post last spring.
There were a lot of opinions about TIMEIT expressed in the comments of that blog post, including some of mine.
My blog post today gives a more full history of the function, its design goals, and how it works. I thought it might prompt more discussion, so I'm creating this thread as a place for it.
If you are an interested user of TIMEIT, feel free to weigh in here with your thoughts. Perhaps the thread will influence MathWorks regarding what to do with TIMEIT, or with related performance measurement capabilities.
Hi
If you have used the playground and are familiar with its capabilities, I will be very interested in your opinion about the tool.
Thank you in advance for your reply/opinion.
At
Hi everyone
The R2025a pre-release is now available to licensed users. I highly encourage you to download, give it a try and give us some feedback.
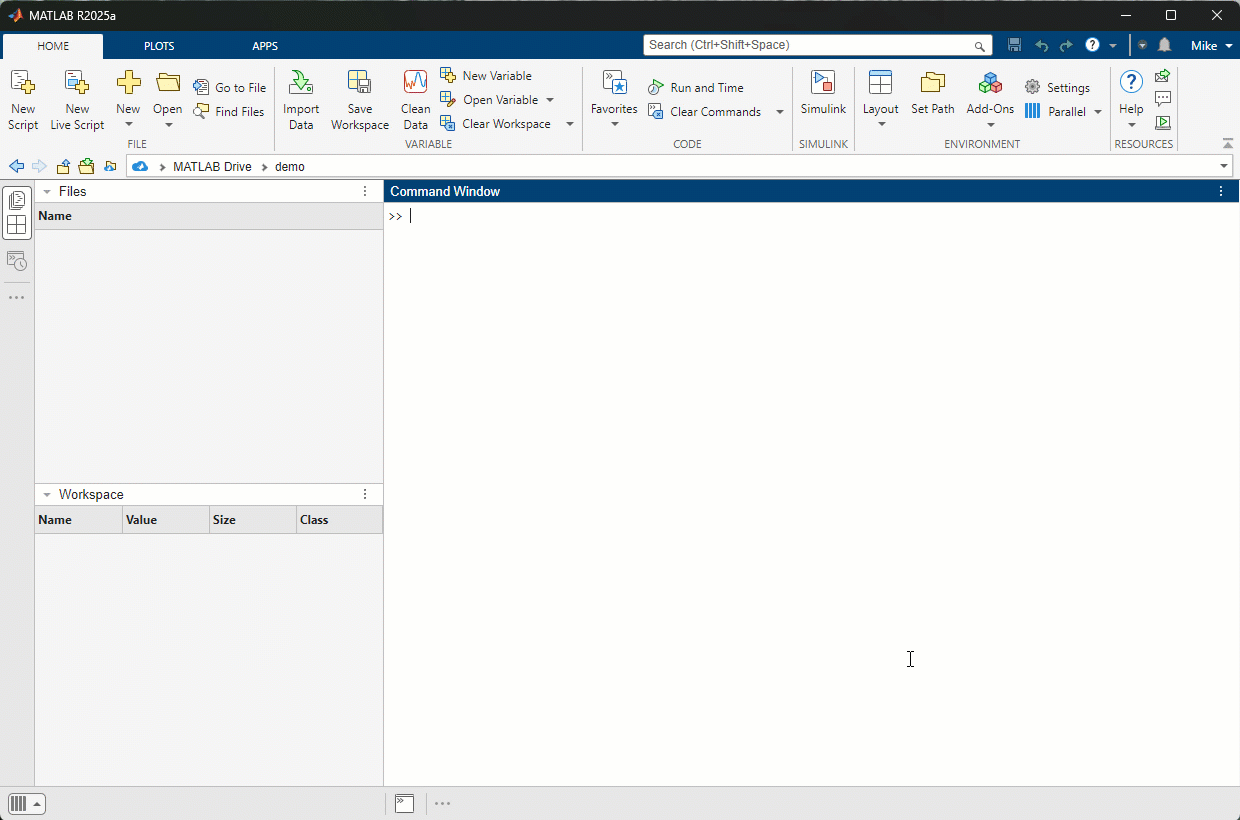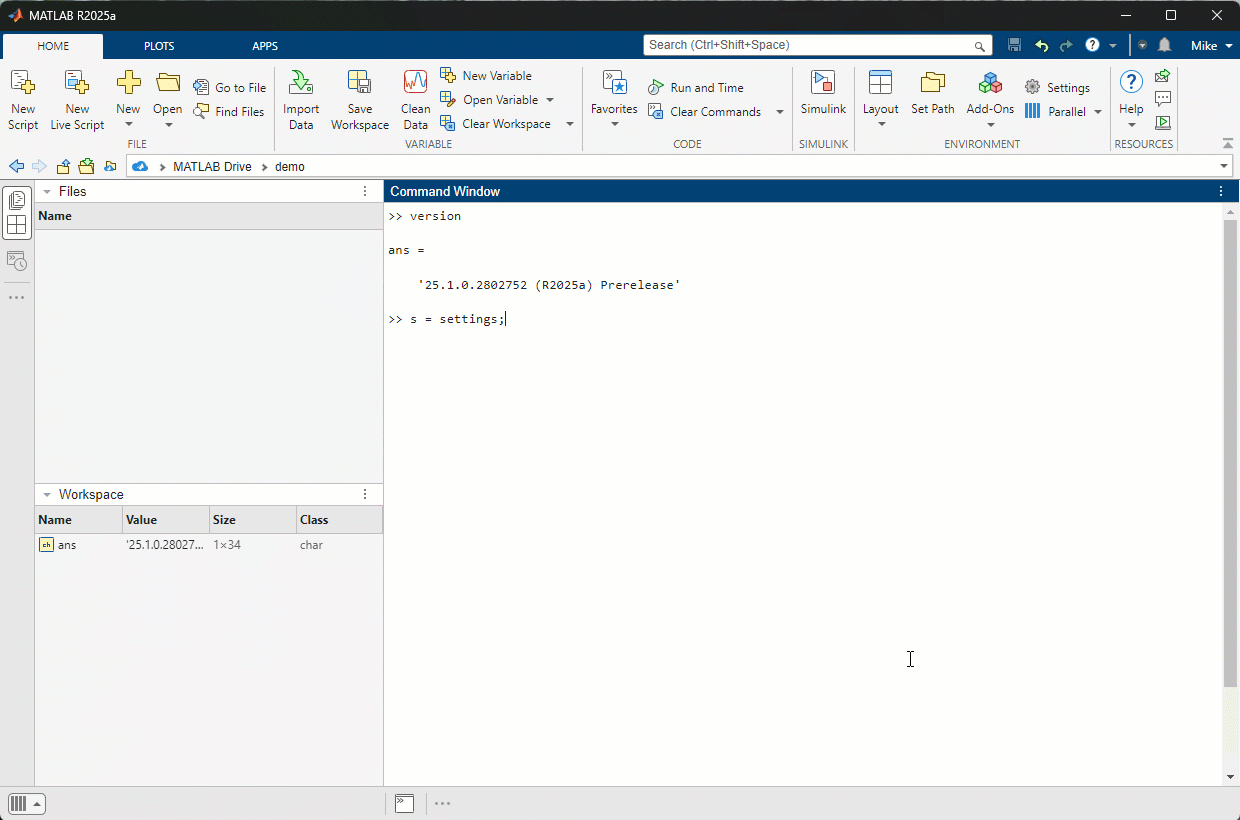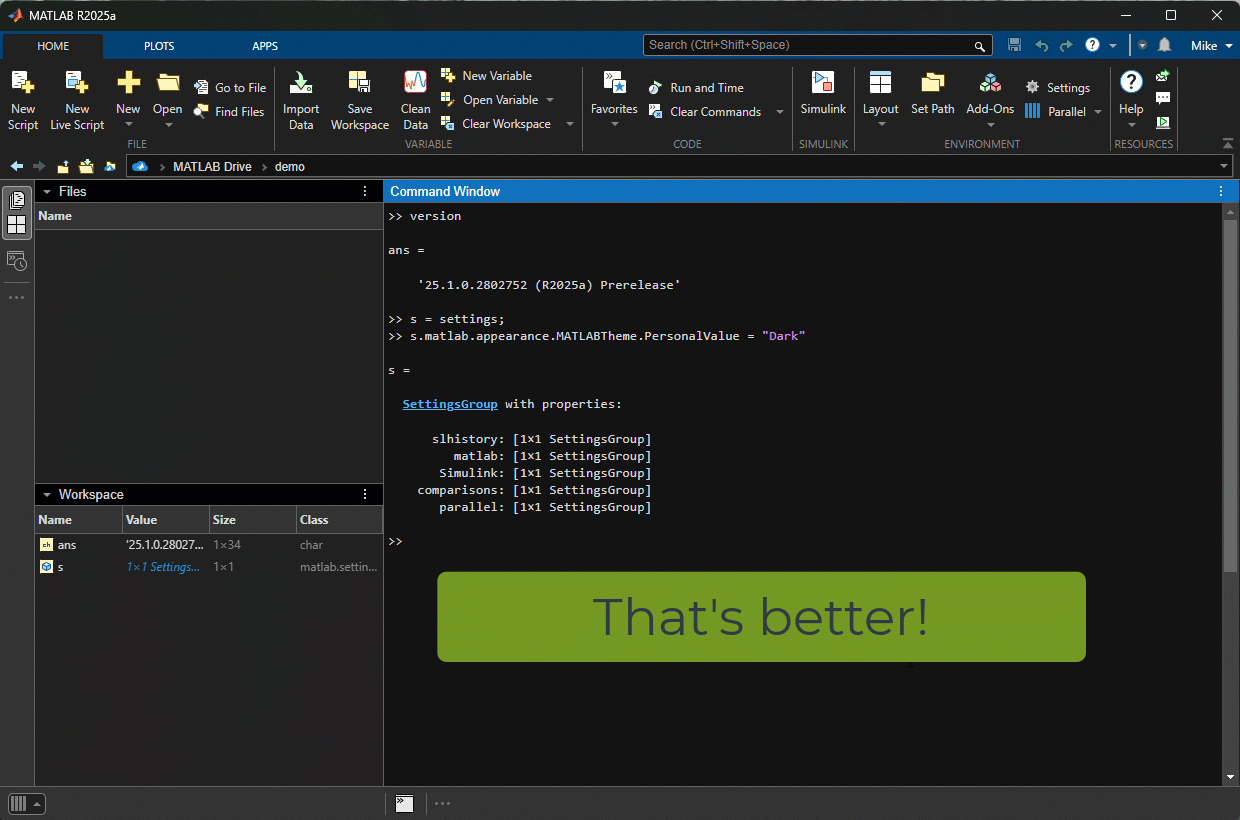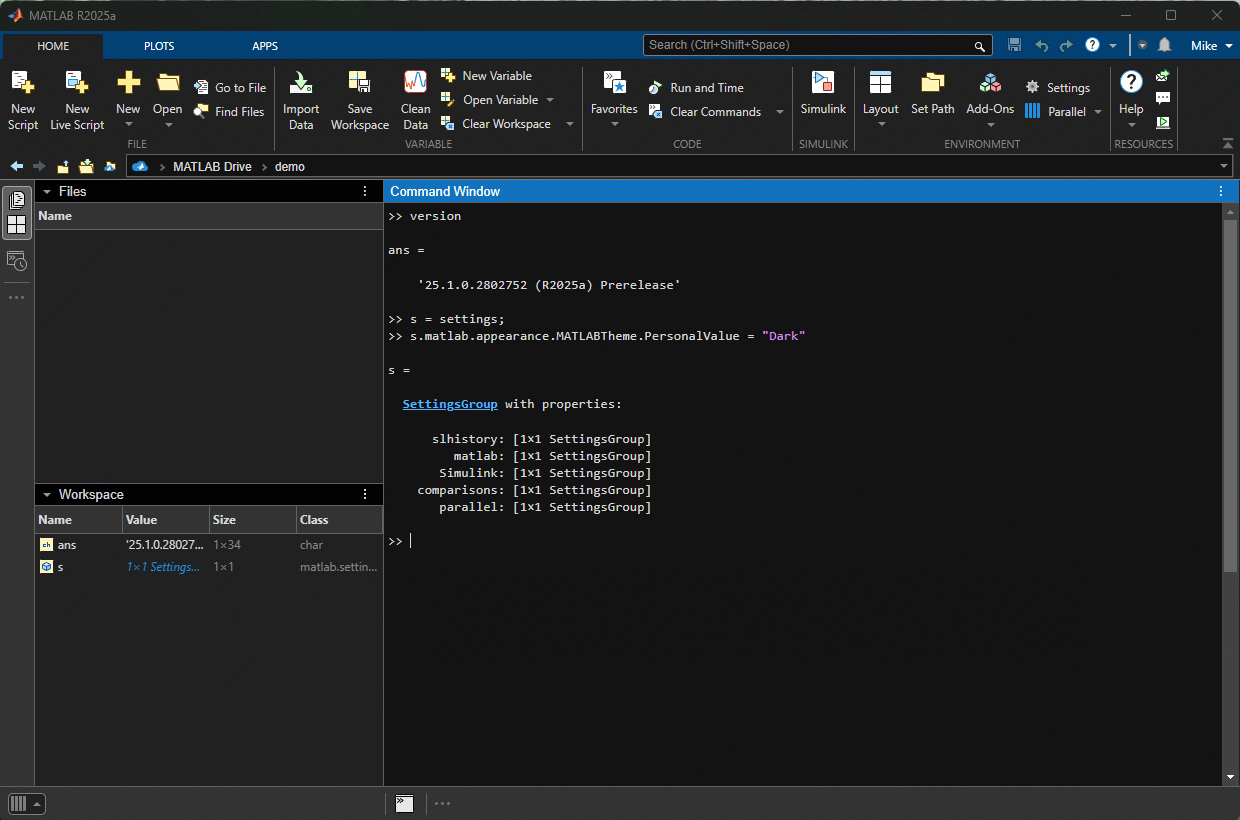
The first thing I tried was switching to Dark mode. Here's the magic
>> s = settings;
>> s.matlab.appearance.MATLABTheme.PersonalValue = "Dark";
The Most Accepted Badge and Top Downloads Badge are two prestigious annual honors that recognize outstanding contributions in MATLAB Answers and File Exchange.
Most Accepted badge is awarded to the top 10 contributors whose answers received the most acceptances. Top Downloads badge goes to the top 10 contributors with the highest number of downloads for their submissions. Please note that, starting in 2025, the criteria for Top Downloaded Badge in will be adjusted to only count downloads from files created or updated in 2025.


In 2024, the recipients for Most Accepted are: @Voss, @Walter Roberson, @Star Strider, @Torsten, @Matt J, @Stephen23, @Steven Lord, @Hassaan, @Sam Chak, and @Cris LaPierre.
The recipients for Top Downloaded are: @Steve Miller, @Rodney Tan, @Yair Altman, @Chad Greene, @William Thielicke, @Seyedali Mirjalili, @John D'Errico, @Giampiero Campa, @Toshiaki Takeuchi, and @Zhaoxu Liu / slandarer.
Congratulations and thank you again for your outstanding contributions in 2024!
Let's celebrate what made 2024 memorable! Together, we made big impacts, hosted exciting events, and built new apps.


Resource links:
Hello,
Now that the "Copilot+PC" (Windows ARM) laptops are rapidly increasing in market share (Microsoft Surface Laptop, Dell XPS 13, HP OmniBook X 14, and more), are there any plans to provide builds for Matlab on Windows arm64?
Since there are already Windows builds of Matlab, it shouldn't be too hard to compile for Windows arm64, as far as I know. But I am not famaliar with Matlab's codebase.
Please try to publish Windows arm64 builds soon so that Matlab can be much more usable on Windows on ARM as it will run natively instead of in emulation.
Thank you very much.
Check out this 3D chart that won Visual Of The Year for 2024 by Visual Capitalist. It's a mashup between a 3D bubblechart and a categorical bar plot yet the only graphical components are the x-axis labels and the legend. Not only does it show relative proportions of material in a laptop but it also shows what the raw material looks like.
I love the idea of analog data visualization. I wonder if any readers have made a analog "chart".
We’d like to announce a change on the Machine Translation feature on MATLAB Answers.
When users are visiting our international domains (e.g. China or Japan), Answers provides the option to translate the content. Recently, we identified several security threats involving high-volume requests from certain IP addresses targeting our translation service.
As one of the countermeasures, we have now placed the Machine Translation feature behind a login requirement. While non-logged-in users will still see the 'Translate' button, it will be inactive (greyed out) until they log in.
We are actively collaborating with adjacent teams to develop solutions to better detect and block malicious requests.
Please let us know if you have any questions or concerns.
The MATLAB Online Training Suite has been updated in the areas of Deep Learning and traditional Machine Learning! These are great self-paced courses that can get you from zero to hero pretty quickly.
Deep Learning Onramp (Free to everyone!) has been updated to use the dlnetwork workflow, such as the trainnet function, which became the preferred method for creating and training deep networks in R2024a.
- Content streamlined to reduce the focus on data processing and feature extraction, and emphasize the machine learning workflow.
- Course example simplified by using a sample of the original data.
- Classification Learner used in the course where appropriate.
The rest of the updates are for subscribers to the full Online Training Suite
The Deep Learning Techniques in MATLAB for Image Applications learning path teaches skills to tackle a variety of image applications. It is made up of the following four short courses:
- Explore Convolutional Neural Networks
- Tune Deep Learning Training Options
- Regression with Deep Learning
- Object Detection with Deep Learning
Two more deep learning short courses are also available:
The Machine Learning Techniques in MATLAB learning path helps learners build their traditional machine learning skill set:
I'm beginning this MATLAB-based numerical methods class, and as I was thinking back to my previous MATLAB/Simulink classes, I definitely remember some projects more fondly than others. One of my most memorable was where I had to use MATLAB to analyze electrocardiogram (ECG) peaks. What about you guys? What are some of the best (or worst 🤭) MATLAB projects or assignments you've been given in the past?
Speaking as someone with 31+ years of experience developing and using imshow, I want to advocate for retiring and replacing it.
The function imshow has behaviors and defaults that were appropriate for the MATLAB and computer monitors of the 1990s, but which are not the best choice for most image display situations in today's MATLAB. Also, the 31 years have not been kind to the imshow code base. It is a glitchy, hard-to-maintain monster.
My new File Exchange function, imview, illustrates the kind of changes that I think should be made. The function imview is a much better MATLAB graphics citizen and produces higher quality image display by default, and it dispenses with the whole fraught business of trying to resize the containing figure. Although this is an initial release that does not yet support all the useful options that imshow does, it does enough that I am prepared to stop using imshow in my own work.
The Image Processing Toolbox team has just introduced in R2024b a new image viewer called imageshow, but that image viewer is created in a special-purpose window. It does not satisfy the need for an image display function that works well with the axes and figure objects of the traditional MATLAB graphics system.
I have published a blog post today that describes all this in more detail. I'd be interested to hear what other people think.
Note: Yes, I know there is an Image Processing Toolbox function called imview. That one is a stub for an old toolbox capability that was removed something like 15+ years ago. The only thing the toolbox imview function does now is call error. I have just submitted a support request to MathWorks to remove this old stub.
The int function in the Symbolic Toolbox has a hold/release functionality wherein the expression can be held to delay evaluation
syms x I
eqn = I == int(x,x,'Hold',true)
which allows one to show the integral, and then use release to show the result
release(eqn)
Maybe it would be nice to be able to hold/release any symbolic expression to delay the engine from doing evaluations/simplifications that it typically does. For example:
x*(x+1)/x, sin(sym(pi)/3)
If I'm trying to show a sequence of steps to develop a result, maybe I want to explicitly keep the x/x in the first case and then say "now the x in the numerator and denominator cancel and the result is ..." followed by the release command to get the final result.
Perhaps held expressions could even be nested to show a sequence of results upon subsequent releases.
Held expressions might be subject to other limitations, like maybe they can't be fplotted.
Seems like such a capability might not be useful for problem solving, but might be useful for exposition, instruction, etc.
We will be updating the MATLAB Answers infrastructure at 1PM EST today. We do not expect any disruption of service during this time. However, if you notice any issues, please be patient and try again later. Thank you for your understanding.
Always and almost immediately!
26%
Never
30%
After validating existing code
15%
Y'all get the new releases?
29%
1843 个投票
Many of my best friends at MathWorks speak Spanish as their first language and we have a large community of Spanish-speaking users. You can see good evidence of this by checking out our relatively new Spanish YouTube channel which recently crossed the 10,000 subscriber mark
I've always used MATLAB with other languages. In the early days, C and C++ via mex files were the most common ways I spliced two languages together. Other than that I've also used MATLAB with Java, Excel and even Fortran.
In more recent years, Python is the language I tend to use most alongside MATLAB and support for this combination is steadily improving. In my latest blog post, I show how easy it has become to use Python's Numpy with MATLAB.
Have you used this functionality much? If so, what for? How well did it work for you?
I am inspired by the latest video from YouTube science content creator Veritasium on his distinct yet thorough explanation on how rainbows work. In his video, he set up a glass sphere experiment representing how light rays would travel inside a raindrop that ultimately forms the rainbow. I highly recommend checking it out.
In the meantime, I created an interactive MATLAB App in MATLAB Online using App Designer to visualize the light paths going through a spherical raindrop with numerical calculations along the way. While I've seen many diagrams out there showing the light paths, I haven't found any doing calculations in each step. Hence I created an app in MATLAB to show the calculations along with the visualizations as one varies the position of the incoming light ray.
Demo video:
For more information about the app and how to open it and play around with it in MATLAB Online, please check out my blog article:

