Modeling Biology with SimBiology: An Introduction for iGEM Teams
View the recorded webinar here:
http://www.mathworks.com/videos/modeling-biology-with-simbiology-an-introduction-for-igem-teams-81817.html
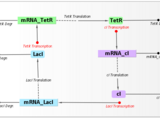
In this webinar, we introduced iGEM teams to SimBiology, a MATLAB-based tool for modeling, simulating and analyzing biological systems. Using the Repressilator [1] model as an example, we demonstrated key features:
- Graphical modeling environment
- Ordinary differential equations (ODEs) and stochastic solvers
- Sensitivity analysis and parameter sweeps techniques
- Parameter estimation techniques
- Advanced analysis via custom analysis tasks written in MATLAB
Reference:
[1] Michael B. Elowitz and Stanislas Leibler , A Synthetic Oscillatory Network of Transcriptional Regulators, Nature. 2000. Jan 20, 403(6767):335-8.
引用格式
Fulden Buyukozturk (2025). Modeling Biology with SimBiology: An Introduction for iGEM Teams (https://ww2.mathworks.cn/matlabcentral/fileexchange/31849-modeling-biology-with-simbiology-an-introduction-for-igem-teams), MATLAB Central File Exchange. 检索时间: .
MATLAB 版本兼容性
平台兼容性
Windows macOS Linux类别
标签
社区
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!