癌症检测
此示例说明如何训练一个神经网络来使用蛋白质表达谱上的质谱数据检测癌症。
简介
血清蛋白质组模式诊断可用于区分疾病患者和非疾病患者的样本。表达谱模式使用表面增强激光解吸和电离 (SELDI) 蛋白质质谱生成。此技术有可能改进癌症病理学临床诊断测试。
问题:癌症检测
目标是构建一个可根据质谱数据区分癌症患者和对照患者的分类器。
此示例中遵循的方法是选择一组精简测量值或“特征”,这些测量值或“特征”可用于通过分类器区分癌症患者和对照患者。这些特征是特定质量/电荷值下的离子强度水平。
整理数据
此示例中使用的数据在文件 ovarian_dataset.mat 中提供,来自 FDA-NCI 临床蛋白质组学计划数据库。有关此数据集的详细说明,请参阅 [1] 和 [2]。
按照Batch Processing of Spectra Using Sequential and Parallel Computing (Bioinformatics Toolbox)中的步骤创建数据文件 OvarianCancerQAQCdataset.mat。新文件包含变量 Y、MZ 和 grp。
Y 中的每列表示从一名患者身上获取的测量值。Y 中有 216 列,表示有 216 个患者,其中 121 个是卵巢癌患者,95 个是非癌症患者。
Y 中的每行表示 MZ 中指示的特定质量-电荷值下的离子强度水平。MZ 中有 15000 个质量-电荷值,Y 中的每行代表在特定质量-电荷值下患者的离子强度水平。
变量 grp 保存关于哪些样本表示癌症患者以及哪些样本表示非癌症患者的索引信息。
关键特征排名
此任务是一个典型的分类问题,其中特征的数量远大于观测值的数量,但是单个特征即可实现正确分类。因此,目标是找到一个分类器,该分类器应适当学习如何加权多个特征,同时生成不会过拟合的广义映射。
找到重要特征的一个简单方法是,假设每个 M/Z 值都是独立的,并计算双向 t 检验。rankfeatures 返回最重要的 M/Z 值的索引,例如,按检验统计量绝对值排名的 100 个索引。
加载 OvarianCancerQAQCdataset.mat 并使用 rankfeatures (Bioinformatics Toolbox) 对特征进行排名,以选择 100 个排名最高的测量值作为输入 x。
ind = rankfeatures(Y,grp,'Criterion','ttest','NumberOfIndices',100); x = Y(ind,:);
为两个类定义目标 t,如下所示:
t = double(strcmp('Cancer',grp));
t = [t; 1-t];
上面列出的脚本和示例中的预处理步骤旨在演示一组有代表性的可能预处理和特征选择过程。使用不同的步骤或参数会产生不同并且可能更好的结果。
[x,t] = ovarian_dataset; whos x t
Name Size Bytes Class Attributes t 2x216 3456 double x 100x216 172800 double
x 中的每一列表示 216 个不同患者中的一个。
x 中的每行表示每个患者在 100 个特定质量-电荷值之一下的离子强度水平。
变量 t 具有两行,包含 216 个值,其中每个值为 [1;0](表示癌症患者)或 [0;1](表示非癌症患者)。
使用前馈神经网络进行分类
现在您已确定一些重要特征,您可以使用这些信息对癌症样本和正常样本进行分类。
由于神经网络以随机初始权重进行初始化,因此每次运行该示例来训练网络后的结果都略有不同。为了避免这种随机性,请设置随机种子以便每次都重现相同的结果。但设置随机种子对于您自己的应用情形并不是必需的。
setdemorandstream(672880951)
创建并训练具有 5 个隐藏层神经元的单隐藏层前馈神经网络。输入样本和目标样本自动分为训练集、验证集和测试集。训练集用于对网络进行训练。只要网络针对验证集持续改进,训练就会继续。测试集提供独立的网络准确度测量。
输入和输出的大小为 0,因为网络尚未配置成与输入数据和目标数据相匹配。在训练网络时会进行此配置。
net = patternnet(5); view(net)

现在网络已准备就绪,可以开始训练。样本自动分为训练集、验证集和测试集。训练集用于对网络进行训练。只要网络针对验证集持续改进,训练就会继续。测试集提供独立的网络准确度测量。
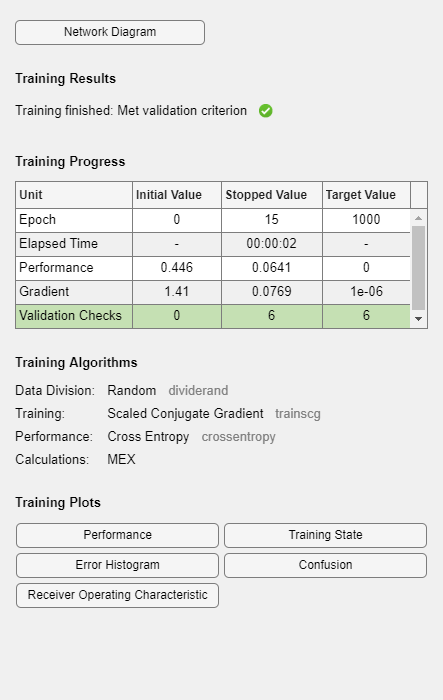
神经网络训练工具显示正在接受训练的网络和用于训练该网络的算法。它还显示训练过程中的训练状态以及停止训练的条件(以绿色突出显示)。
底部的按钮用于打开有用的绘图,这些图可以在训练期间和训练后打开。算法名称和绘图按钮旁边的链接可打开有关这些主题的文档。
[net,tr] = train(net,x,t);
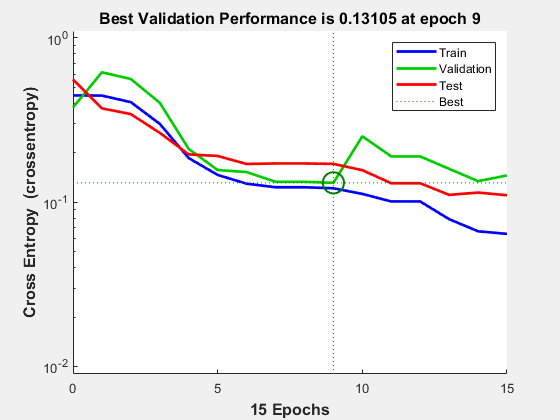
要查看在训练过程中网络性能的改善情况,请点击训练工具中的 Performance 按钮,或使用 plotperform 函数。
性能以均方误差衡量,并以对数刻度显示。随着网络训练的加深,均方误差迅速降低。
绘图会显示训练集、验证集和测试集的性能。
plotperform(tr)
现在可以使用我们从主数据集划分出来的测试样本测试经过训练的神经网络。在训练中没有以任何方式使用过测试数据,因此测试数据是可用来测试网络的“样本外”数据集。这样可以估计出当使用真实数据进行测试时,网络的表现如何。
网络输出的范围为 0-1。对输出应用阈值以获得 1 和 0,分别表示癌症患者和非癌症患者。
testX = x(:,tr.testInd); testT = t(:,tr.testInd); testY = net(testX); testClasses = testY > 0.5
testClasses = 2×32 logical array
1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 1 1 0 0 1 1 0 0 0 0 0 0 0 0 0 1 0
0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 1 1 0 0 1 1 1 1 1 1 1 1 1 0 1
衡量神经网络数据拟合程度的一个方法是混淆矩阵图。
该混淆矩阵显示了正确和错误分类的百分比。正确分类表示为矩阵对角线上的绿色方块。红色方块表示错误分类。
如果网络是准确的,则红色方块中的百分比应该很小,表示几乎没有错误分类。
如果网络不准确,则您可以尝试训练更长时间,或者训练具有更多隐藏神经元的网络。
plotconfusion(testT,testY)
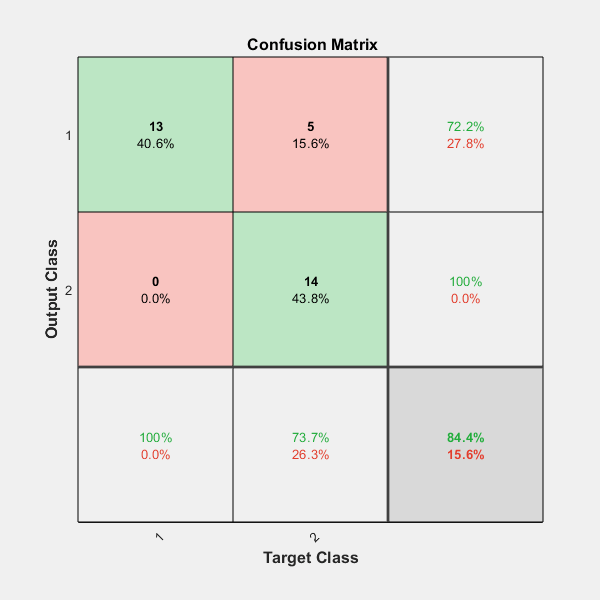
以下是正确和错误分类的总体百分比。
[c,cm] = confusion(testT,testY);
fprintf('Percentage Correct Classification : %f%%\n', 100*(1-c));Percentage Correct Classification : 84.375000%
fprintf('Percentage Incorrect Classification : %f%%\n', 100*c);Percentage Incorrect Classification : 15.625000%
衡量神经网络数据拟合程度的另一个方法是受试者工作特征图。该图显示随着输出阈值从 0 变为 1,假正率和真正率之间的相关性。
线条越偏向左上方,达到高的真正率所需接受的假正数越少。最佳分类器是线条从左下角到左上角再到右上角,或接近于该模式。
第 1 类表示癌症患者,第 2 类表示非癌症患者。
plotroc(testT,testY)

此示例演示了如何将神经网络用作癌症检测的分类器。为了提高分类器性能,您还可以尝试使用主成分分析等技术来降低用于神经网络训练的数据的维度。
参考资料
[1] T.P. Conrads, et al., "High-resolution serum proteomic features for ovarian detection", Endocrine-Related Cancer, 11, 2004, pp. 163-178.
[2] E.F. Petricoin, et al., "Use of proteomic patterns in serum to identify ovarian cancer", Lancet, 359(9306), 2002, pp. 572-577.