指定自定义权重初始化函数
此示例说明如何为后跟泄漏 ReLU 层的卷积层创建自定义 He 权重初始化函数。
这个 He 初始化函数从均值为零、方差为 的正态分布进行采样,其中 a 是卷积层后的泄漏 ReLU 层的缩放因子,n = FilterSize(1) * FilterSize(2) * NumChannels。
对于可学习层,当选项 WeightsInititializer、InputWeightsInitializer 或 RecurrentWeightsInitializer 设置为 "he" 时,软件使用 a=0。要将 a 设置为不同值,请创建一个自定义函数以用作权重初始化函数。
加载数据
将数字样本数据加载为图像数据存储。imageDatastore 函数根据文件夹名称自动对图像加标签。
unzip("DigitsData.zip"); imds = imageDatastore("DigitsData", ... IncludeSubfolders=true, ... LabelSource="foldernames");
将数据划分为训练数据集和验证数据集,以使训练集中的每个类别包含 750 个图像,并且验证集包含对应每个标签的其余图像。splitEachLabel 将数据存储拆分为两个新的数据存储以用于训练和验证。
numTrainFiles = 750;
[imdsTrain,imdsValidation] = splitEachLabel(imds,numTrainFiles,"randomize");创建自定义权重初始化函数
leakyHe 函数接受输入 sz(层权重的大小),并返回后跟泄漏 ReLU 层的卷积层的 He 初始化函数给出的权重数组。该函数还接受可选输入参量 scale,该参量指定泄漏 ReLU 层的尺度乘数。
function weights = leakyHe(sz,scale) % If not specified, then use default scale = 0.1 if nargin < 2 scale = 0.1; end filterSize = [sz(1) sz(2)]; numChannels = sz(3); numIn = filterSize(1) * filterSize(2) * numChannels; varWeights = 2 / ((1 + scale^2) * numIn); weights = randn(sz) * sqrt(varWeights); end
定义网络架构
定义卷积神经网络架构:
大小为
[28 28 1]的图像输入层,输入图像的大小三个二维卷积层,滤波器大小为 3 并分别具有 8、16 和 32 个滤波器
每个卷积层后有一个泄漏 ReLU 层
大小为 10 的全连接层,类的数量
Softmax 层
对于每个卷积层,将权重初始化函数设置为 leakyHe 函数。
inputSize = [28 28 1];
numClasses = 10;
layers = [
imageInputLayer(inputSize)
convolution2dLayer(3,8,WeightsInitializer=@leakyHe)
leakyReluLayer
convolution2dLayer(3,16,WeightsInitializer=@leakyHe)
leakyReluLayer
convolution2dLayer(3,32,WeightsInitializer=@leakyHe)
leakyReluLayer
fullyConnectedLayer(numClasses)
softmaxLayer];训练网络
指定训练选项并训练网络。进行四轮训练。要防止梯度爆炸,请将梯度阈值设置为 2。每轮训练后对网络进行一次验证。在图中显示训练进度并监控准确度。
使用 trainnet 函数训练神经网络。对于分类,使用交叉熵损失。默认情况下,trainnet 函数使用 GPU(如果有)。在 GPU 上进行训练需要 Parallel Computing Toolbox™ 许可证和受支持的 GPU 设备。有关受支持设备的信息,请参阅GPU 计算要求 (Parallel Computing Toolbox)。否则,trainnet 函数使用 CPU。要指定执行环境,请使用 ExecutionEnvironment 训练选项。
maxEpochs = 4; miniBatchSize = 128; numObservations = numel(imdsTrain.Files); numIterationsPerEpoch = floor(numObservations / miniBatchSize); options = trainingOptions("sgdm", ... MaxEpochs=maxEpochs, ... MiniBatchSize=miniBatchSize, ... GradientThreshold=2, ... ValidationData=imdsValidation, ... ValidationFrequency=numIterationsPerEpoch, ... Verbose=false, ... Metrics="accuracy", ... Plots="training-progress"); [netDefault,infoDefault] = trainnet(imdsTrain,layers,"crossentropy",options);

测试网络
使用 testnet 函数测试神经网络。对于单标签分类,需评估准确度。准确度是指正确预测的百分比。默认情况下,testnet 函数使用 GPU(如果有)。要手动选择执行环境,请使用 testnet 函数的 ExecutionEnvironment 参量。
accuracy = testnet(netDefault,imdsValidation,"accuracy");指定附加选项
leakyHe 函数接受可选输入参量 scale。要将额外的变量输入到自定义权重初始化函数中,请将该函数指定为接受单个输入 sz 的匿名函数。为此,请用 @(sz)leakyHe(sz,scale) 替换 @leakyHe 的实例。此处,匿名函数只接受单个输入参量 sz,并使用指定的 scale 输入参量调用 leakyHe 函数。
对网络进行以下更改,然后沿用前面的方法训练网络:
对于泄漏 ReLU 层,指定 0.01 的尺度乘数。
使用
leakyHe函数初始化卷积层的权重,并指定尺度乘数。
scale = 0.01;
layers = [
imageInputLayer(inputSize)
convolution2dLayer(3,8,WeightsInitializer=@(sz)leakyHe(sz,scale))
leakyReluLayer(scale)
convolution2dLayer(3,16,WeightsInitializer=@(sz)leakyHe(sz,scale))
leakyReluLayer(scale)
convolution2dLayer(3,32,WeightsInitializer=@(sz)leakyHe(sz,scale))
leakyReluLayer(scale)
fullyConnectedLayer(numClasses)
softmaxLayer];
[netCustom,infoCustom] = trainnet(imdsTrain,layers,"crossentropy",options);
使用 testnet 函数测试网络。
accuracy = testnet(netCustom,imdsValidation,"accuracy");比较结果
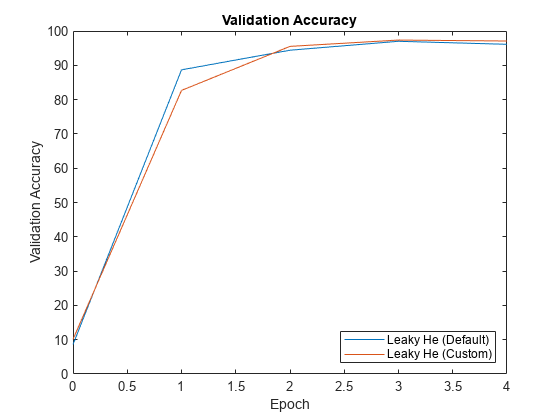
从 trainnet 函数的信息输出中提取验证准确度。
validationAccuracy = [
infoDefault.ValidationHistory.Accuracy, ...
infoCustom.ValidationHistory.Accuracy];对于每个网络,绘制轮次编号对验证准确度的图。
figure epochs = 0:maxEpochs; plot(epochs,validationAccuracy) title("Validation Accuracy") xlabel("Epoch") ylabel("Validation Accuracy") legend(["Leaky He (Default)" "Leaky He (Custom)"],'Location','southeast')
参考书目
He, Kaiming, Xiangyu Zhang, Shaoqing Ren, and Jian Sun."Delving deep into rectifiers:Surpassing human-level performance on imagenet classification."In Proceedings of the IEEE international conference on computer vision, pp. 1026-1034. 2015.
另请参阅
trainnet | trainingOptions | dlnetwork