imclearborder
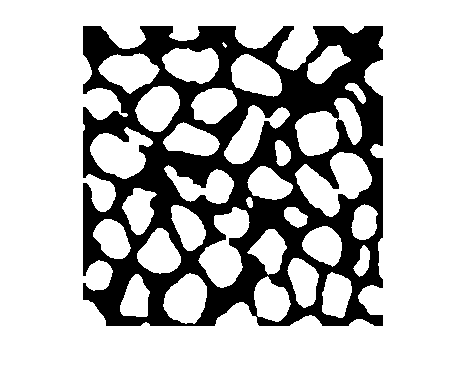
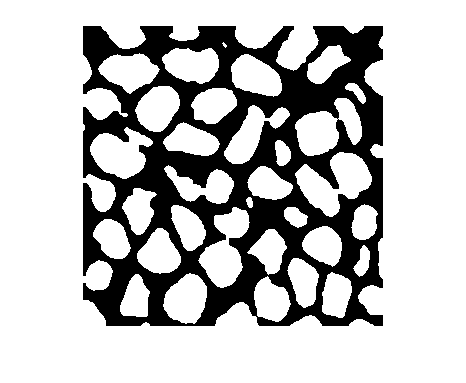
隐藏连接到图像边界的光亮结构
说明
J = imclearborder(___,Name=Value)imclearborder(I,Borders=["left" "right"]) 仅删除触及图像左边界或右边界的结构。
示例
输入参数
名称-值参数
输出参量
算法
imclearborder 使用形态学重新构造,其中:
掩膜图像是输入图像。
标记图像边界上的元素等于掩膜图像,其他位置的元素都为 0。
参考
[1] Soille, Pierre. Morphological Image Analysis: Principles and Applications Berlin ; New York: Springer, 1999, 164–165.
[2] Molnar, Ian. "Uniform Quartz - Silver Nanoparticle Injection Experiment." Digital Rocks Portal (April 2025). Accessed August 8, 2025. https://www.doi.org/10.17612/P7Z59J.