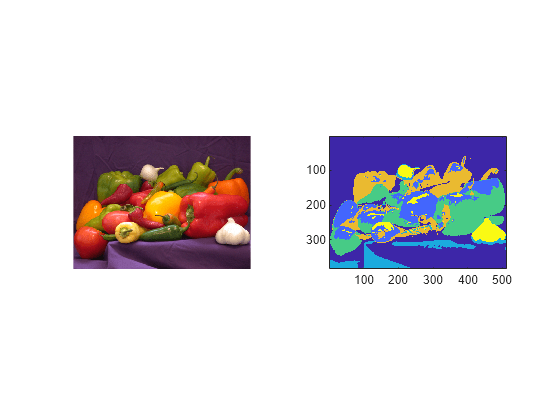
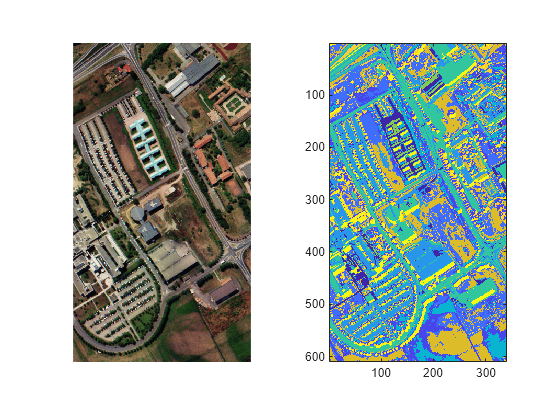
imsegisodata
Description
The iterative self-organizing data analysis technique (ISODATA) algorithm is a
method used in pattern recognition and image analysis for clustering multivariate data into
naturally occurring groups. Developed as an extension of the k-means
clustering algorithm, ISODATA clusters data by iteratively adjusting clusters and
automatically determining their optimal number. Compared to the k-means
image segmentation of the imsegkmeans
function, the ISODATA image segmentation of imsegisodata is more flexible
and powerful, particularly in handling real-world, complex, and varied data sets, where the
number of clusters is not already known.
[
fine-tunes the ISODATA algorithm using one or more optional name-value arguments. For
example, L,centers] = imsegisodata(I,Name=Value)InitialNumClusters=5 starts the clustering process with five
clusters.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
The imsegisodata function segments the image I using
these steps, and returns the assigned cluster labels L and cluster centers
centers.
Normalization — You can choose to normalize the input image, if the channels of the image have different intensity ranges, by specifying the
NormalizeInputname-value argument astrue.Initialization — The function starts with an initial set of cluster centers, which you can select randomly or based on prior knowledge. The function assigns each pixel to the nearest cluster center based on the Euclidean distance. You can specify the initial number of clusters by using the
InitialNumClustersname-value argument.Update — After the function completes the initial assignment of each pixel to a cluster, it iteratively removes empty clusters, splits clusters, and merges clusters until it reaches a stopping criterion such as the maximum number of iterations specified by
MaxIterations. After each update, the function recalculates each cluster center as the mean of the pixels in that cluster.Remove Small Clusters — The function removes clusters that have fewer than
MinSamplespixels.Splitting Clusters — The function evaluates each cluster to determine whether to split it into two. It splits a cluster into two based on several criteria, such as if the standard deviation of the pixels within the cluster is greater than
MaxStandardDeviation.Merging Clusters — The function evaluates whether to merge any pairs of clusters. It merges two clusters if the distance between their centers is less than or equal to
MinClusterSeparation. The center of the merged cluster is the weighted average of the cluster centers of the two clusters that are merged. The function merges at mostMaxPairsToMergecluster pairs per iteration.
Depending on the parameters specified for the ISODATA algorithm, segmentation
using the imsegisodata function may be time-consuming or may spiral out of
control leaving only one class, if the data is very unstructured.
References
[1] Tou, Julius T., and Rafael C. Gonzalez. Pattern Recognition Principles. Applied Mathematics and Computation, No. 7. Reading, Mass: Addison-Wesley Pub. Co, 1974.