高光谱图像处理
导入、导出、处理和可视化高光谱数据
Hyperspectral Imaging Library for Image Processing Toolbox™ 为高光谱和多光谱图像处理和可视化提供 MATLAB® 函数和工具。
使用此库中的函数可读取、写入和处理通过使用高光谱和多光谱成像传感器以各种文件格式捕获的数据。该库支持国家图像传输格式 (NITF)、可视化图像环境 (ENVI)、标记图像文件格式 (TIFF)、元数据文本扩展 (MTL)、分层数据格式 (HDF) 和欧洲标准存档格式 (SAFE) 文件格式。
该库提供一套算法,用于条带选择、去噪、辐射和大气校正、降维、端元提取、丰度图估计、光谱匹配、异常检测、计算谱指数和分割。
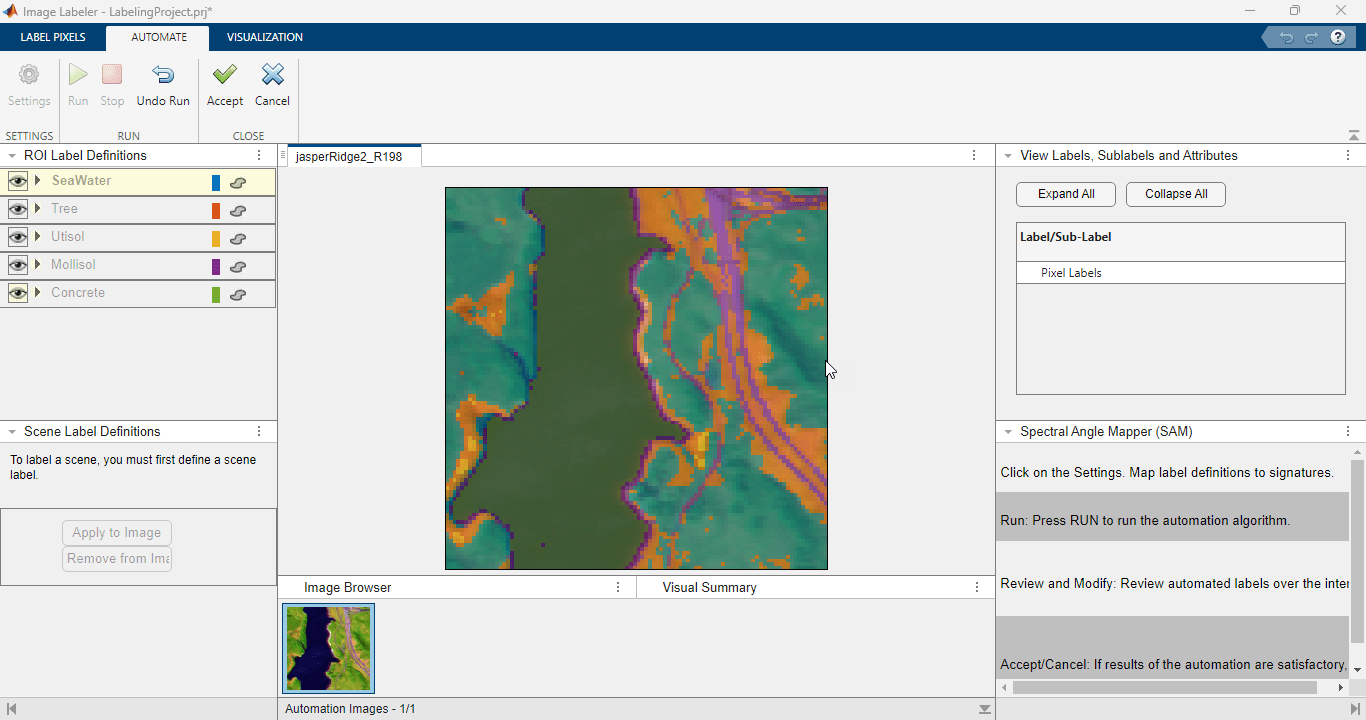
高光谱查看器使您能够读取高光谱和多光谱数据,可视化元数据和地理空间信息,可视化各个条带图像及其直方图,为数据立方体中的一个像素或区域创建一个频谱图,绘制端元,生成不同颜色或假彩色表示,计算谱指数,以及导出结果。
要执行高光谱图像和多光谱图像分析,请从附加功能资源管理器下载 Hyperspectral Imaging Library for Image Processing Toolbox。有关下载附加功能的详细信息,请参阅获取和管理附加功能。
App
| 高光谱查看器 | 可视化高光谱和多光谱数据 |
函数
主题
快速入门
- Get Started with Hyperspectral Image Processing
Basics of hyperspectral image processing. - Analyze Hyperspectral and Multispectral Images
Describes approaches to hyperspectral and multispectral imaging. - Explore Hyperspectral and Multispectral Data in the Hyperspectral Viewer
This example shows how to explore hyperspectral and multispectral data using the 高光谱查看器 app. - Process Large Hyperspectral and Multispectral Images
This example shows how to process small regions of large hyperspectral and multispectral images. - Hyperspectral and Multispectral Data Correction
Describes radiometric calibration, atmospheric correction, and spectral correction. - Spectral Matching and Target Detection Techniques
Techniques for target detection and spectral matching. - Spectral Indices
Describes spectral indices. - Support for Singleton Dimensions
Analysis of 1-D and 2-D spectral data using singleton hypercube.
分类
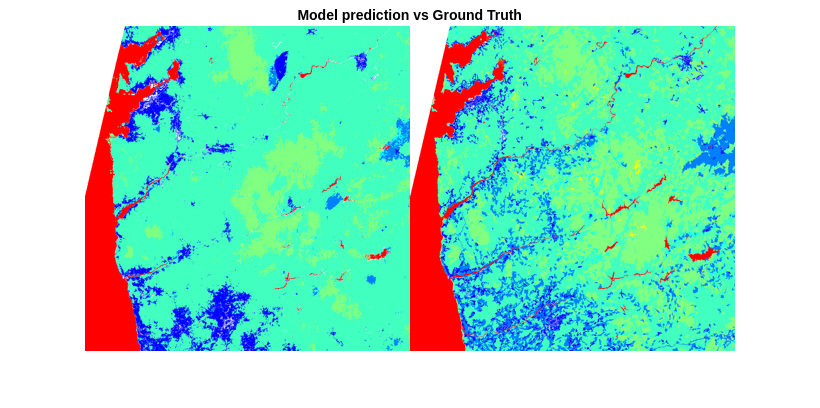
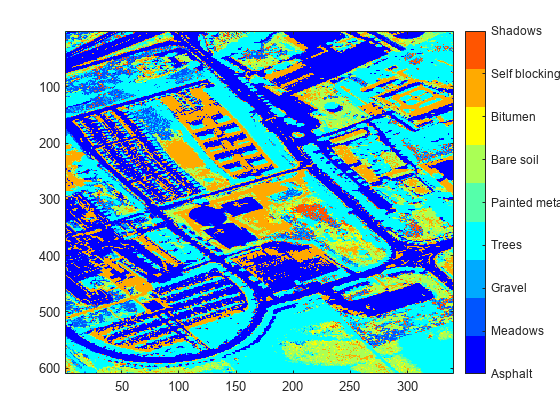
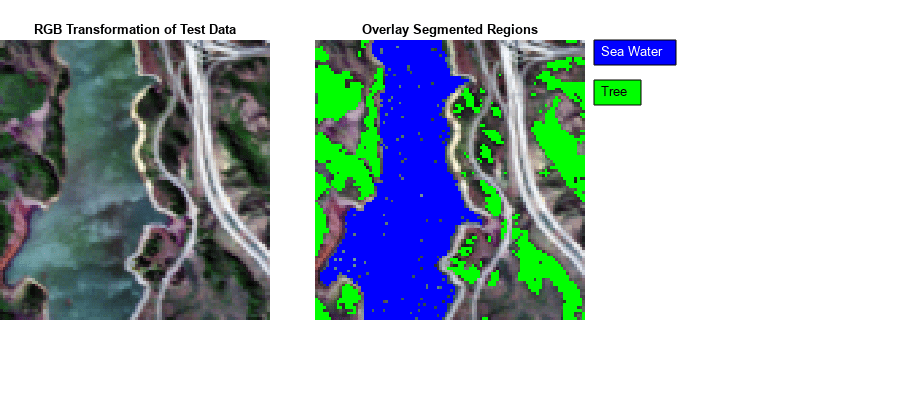
- Classify Hyperspectral Image Using Library Signatures and SAM
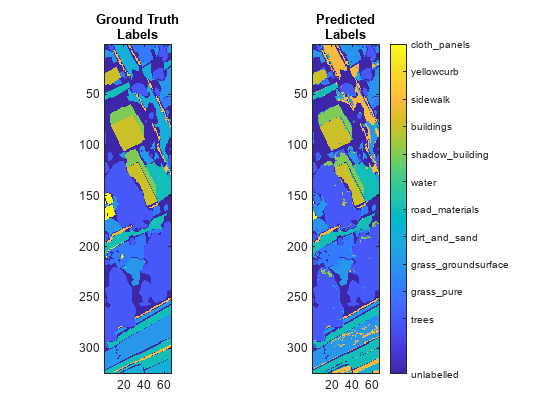
Classify pixels in a hyperspectral image by using the spectral angle mapper (SAM) classification algorithm. - Classify Hyperspectral Images Using Deep Learning
This example shows how to perform hyperspectral image classification using a custom spectral convolution neural network (CSCNN). - Classify Hyperspectral Image Using Support Vector Machine Classifier
This example shows how to perform hyperspectral image classification using a support vector machine (SVM) classifier.
区域标识
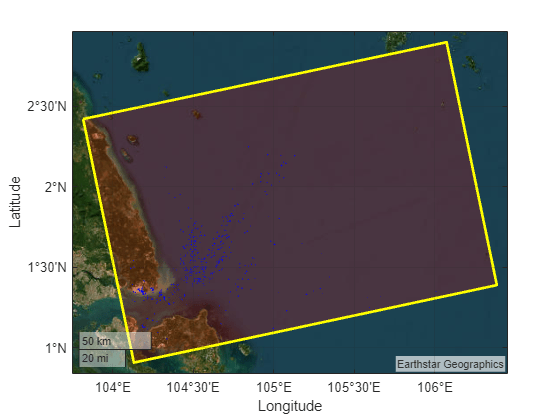
- Target Detection Using Spectral Signature Matching
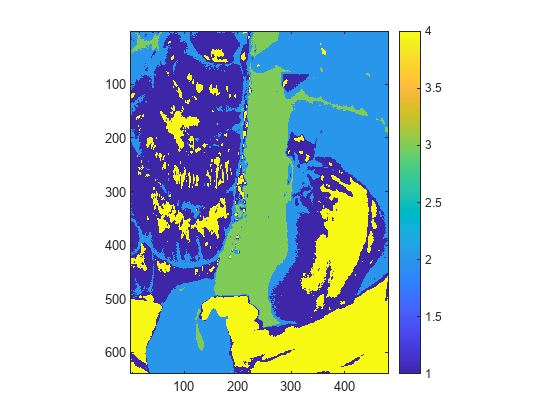
Detect a known target in the hyperspectral image by using the spectral matching method. - Identify Vegetation Regions Using Interactive NDVI Thresholding
Identify the types of vegetations regions in a hyperspectral image through interactive thresholding of a normalized difference vegetation index (NDVI) map. - Find Regions in Spatially Referenced Multispectral Image
This example shows how to identify water and vegetation regions in a Landsat 8 multispectral image and spatially reference the image.
数字孪生
- Generate RoadRunner Scene Using Aerial Hyperspectral and Lidar Data (Automated Driving Toolbox)
Generate RoadRunner scene from aerial hyperspectral and lidar data.
分割
- Interactively Segment Hyperspectral Image Using Segment Anything Model
This example shows how to interactively preprocess and segment a hyperspectral image using the Segment Anything Model (SAM). (自 R2025a 起)