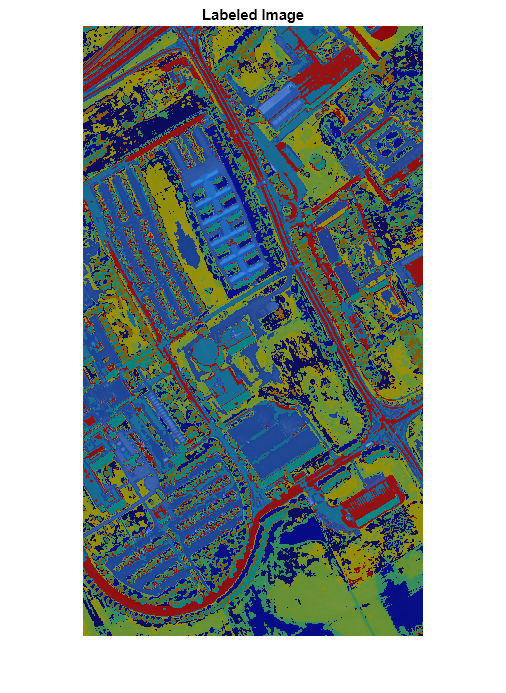
hyperseganchor
Segment hyperspectral and multispectral images using fast spectral clustering with anchor graphs
Since R2024a
Description
You can segment a hyperspectral or multispectral image using both spectral and spatial information with the fast spectral clustering with anchor graph (FSCAG) algorithm.
L = hyperseganchor(spcube,K,Name=Value)MaxIterations=50 specifies to perform a maximum of 50 iterations
in the clustering phase.
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
Algorithms
The FSCAG algorithm constructs an anchor graph between the pixel spectra and the spectra of the anchor points. The algorithm measures the similarity between the pixel spectra and the spectra of the anchor points using both spectral and spatial information, and creates a sparse similarity matrix. The algorithm performs iterative spectral clustering of this similarity matrix to cluster the pixels in the hyperspectral or multispectral image.
References
[1] Wang, Rong, Feiping Nie, and Weizhong Yu. “Fast Spectral Clustering With Anchor Graph for Large Hyperspectral Images.” IEEE Geoscience and Remote Sensing Letters 14, no. 11 (November 2017): 2003–7. https://doi.org/10.1109/LGRS.2017.2746625.
[2] Wang, Cheng-Long, Feiping Nie, Rong Wang, and Xuelong Li. “Revisiting Fast Spectral Clustering with Anchor Graph.” In ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 3902–6. Barcelona, Spain: IEEE, 2020. https://doi.org/10.1109/ICASSP40776.2020.9053271.