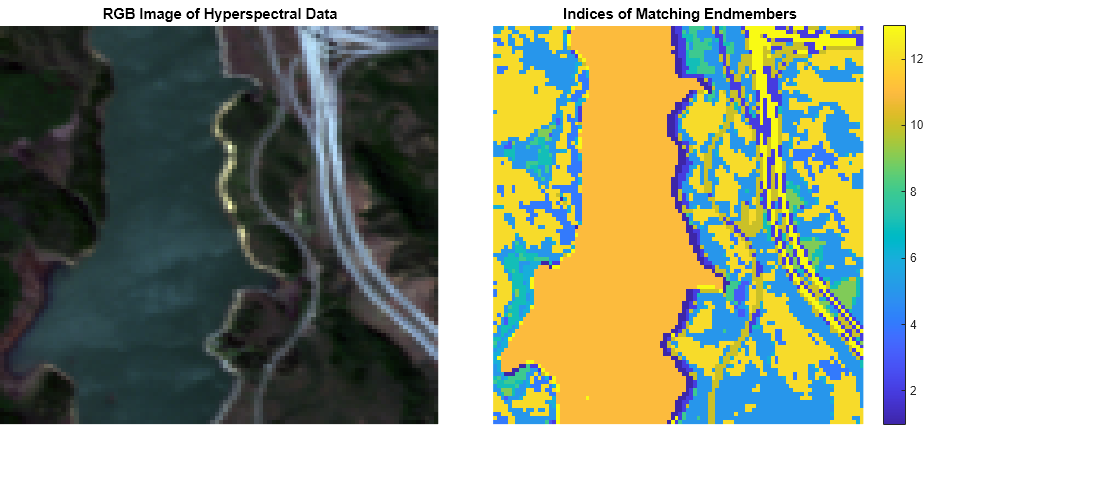
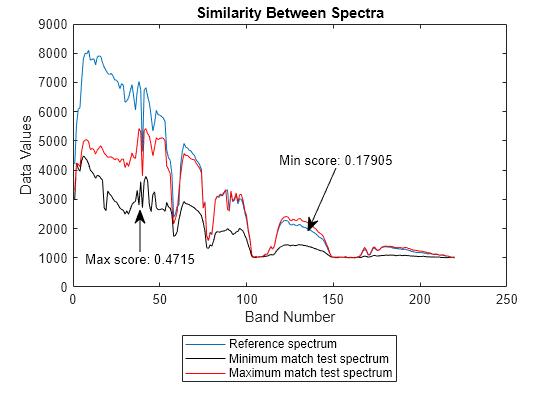
sam
Measure spectral similarity using spectral angle mapper
Description
score = sam(inputData,refSpectra)inputData and the specified reference spectra
refSpectra by using the spectral angle mapper (SAM) classification
algorithm. Use this syntax to identify different regions or materials in a hyperspectral
data cube.
score = sam(testSpectra,refSpectra)testSpectra and reference spectra refSpectra by
using the SAM classification algorithm. Use this syntax to compare the spectral signature of
an unknown material against the reference spectra or to compute spectral variability between
two spectral signatures.
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Examples
Input Arguments
Output Arguments
Limitations
This function does not
support parfor loops, as its performance is already
optimized. (since R2023a)
More About
References
[1] Kruse, F.A., A.B. Lefkoff, J.W. Boardman, K.B. Heidebrecht, A.T. Shapiro, P.J. Barloon, and A.F.H. Goetz. “The Spectral Image Processing System (SIPS)—Interactive Visualization and Analysis of Imaging Spectrometer Data.” Remote Sensing of Environment 44, no. 2–3 (May 1993): 145–63. https://doi.org/10.1016/0034-4257(93)90013-N.
Version History
Introduced in R2020a
See Also
spectralMatch | readEcostressSig | sid | hypercube | sidsam | jmsam