estimateAbundanceLS
Estimate abundance maps
Syntax
Description
abundanceMap = estimateAbundanceLS(inputData,endmembers)
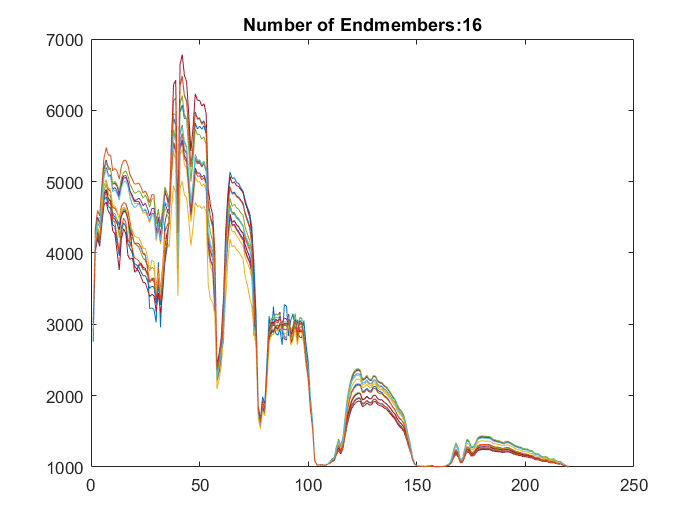
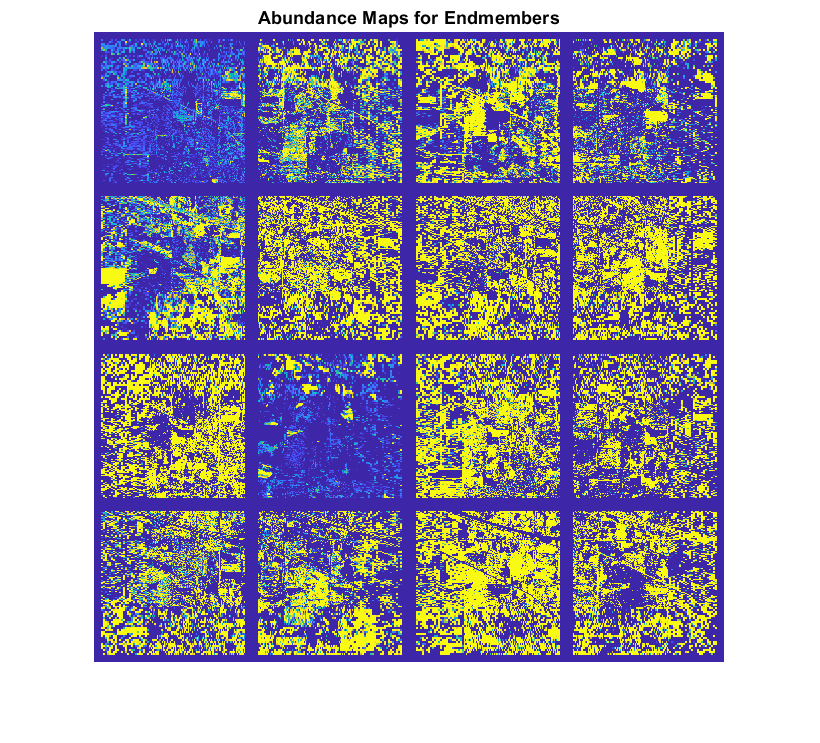
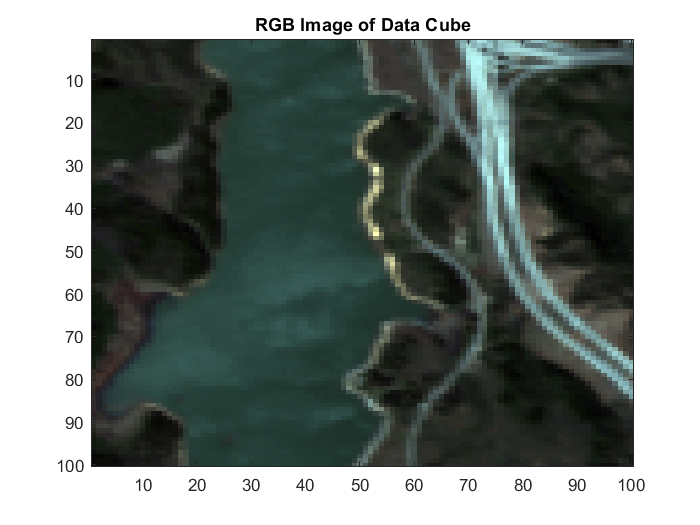
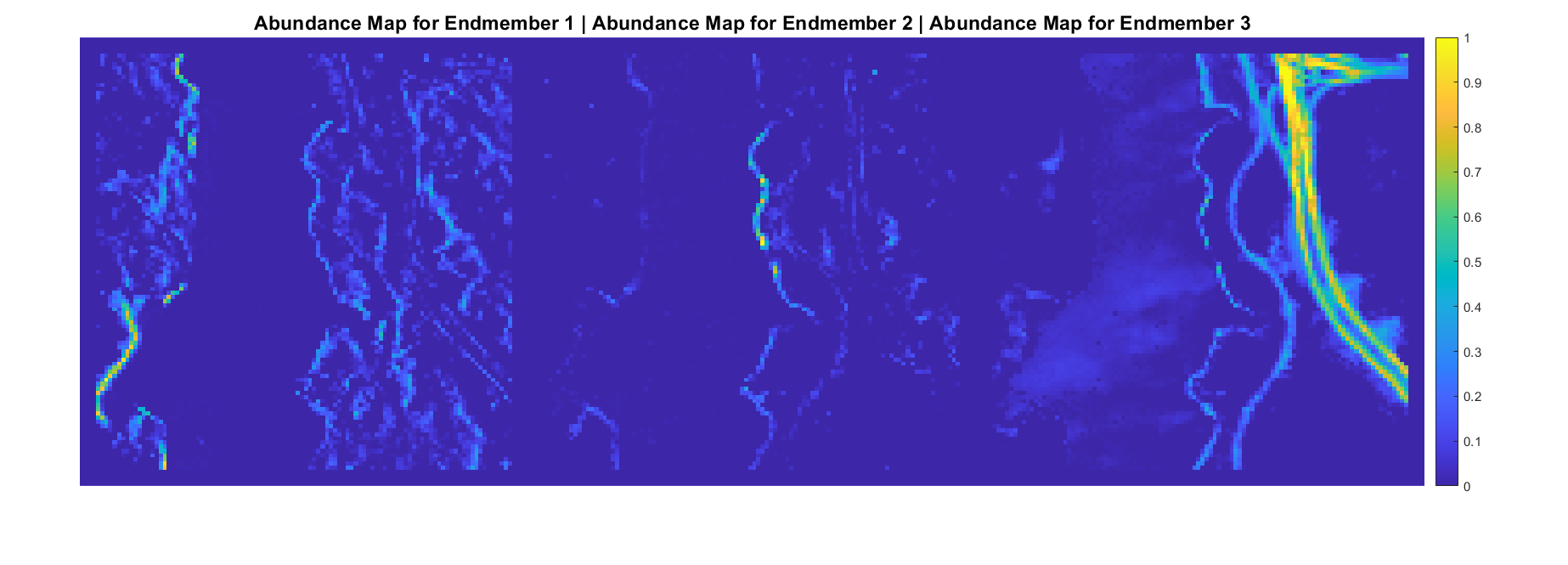
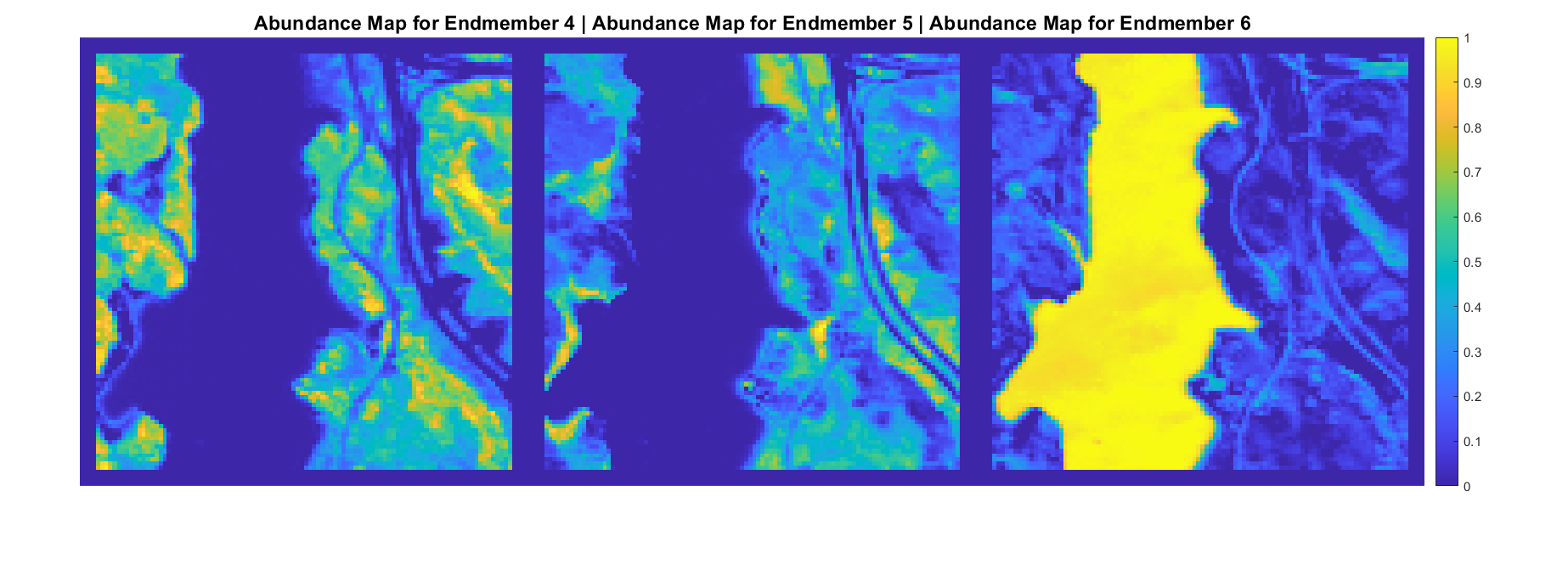
A hyperspectral data cube can contain both pure and mixed pixels. Pure pixels exhibit the spectral characteristics of a single class, while the mixed pixels exhibit the spectral characteristics of multiple classes. The spectral signatures of the pure pixels comprise the endmembers that identify the unique classes present in a hyperspectral data cube. The spectral signature of mixed pixels can be a linear combination of two or more endmember spectra. The abundance map identifies the proportion of each endmember present in the spectra of each pixel. For a hyperspectral data cube of spatial dimensions M-by-N containing P endmembers, there exist P abundance maps, each of size M-by-N.
The abundance map estimation process is known as spectral unmixing, which is the decomposition of the spectra of each pixel into a given set of endmember spectra.
abundanceMap = estimateAbundanceLS(___,Method=estMethod)
Note
This function requires the Hyperspectral Imaging Library for Image Processing Toolbox™. You can install the Hyperspectral Imaging Library for Image Processing Toolbox from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
The Hyperspectral Imaging Library for Image Processing Toolbox requires desktop MATLAB®, as MATLAB Online™ and MATLAB Mobile™ do not support the library.
Examples
Input Arguments
Output Arguments
References
[1] Keshava, N., and J.F. Mustard. “Spectral Unmixing.” IEEE Signal Processing Magazine 19, no. 1 (January 2002): 44–57. https://doi.org/10.1109/79.974727.
[2] Kay, Steven M. Fundamentals of Statistical Signal Processing. Prentice Hall Signal Processing Series. Englewood Cliffs, N.J: Prentice-Hall PTR, 1993.
Version History
Introduced in R2020a