radiomics
Description
The radiomics object and its object functions enable you to
preprocess a medical image of any modality, such as MRI, CT, and ultrasound, and compute shape
features, intensity features, and texture features from a specified region of interest (ROI).
You can use the same set of radiomics features for any medical imaging modality, and for
applications such as studying associations between medical imaging features and patient
biology or predicting clinical outcomes from medical images. The radiomics features computed
using the radiomics object and its object functions comply with the
standards provided by the image biomarker standardisation initiative (IBSI).
Creation
Description
R = radiomics(data,roi)radiomics object for the medical image or volume
data and the region of interest (ROI) roi. You
can compute shape features, intensity features, and texture features in the ROI of the
medical volume using the object. data and roi
set the Data and ROILabel properties, respectively.
R = radiomics(data,roi,PropertyName=Value)
Input Arguments
Medical image or volume to prepare for radiomics, specified as a 2-D numeric
matrix or medicalImage object for a medical image, or a 3-D numeric array or medicalVolume object for a medical volume. This input argument sets the
Data property of the radiomics
object.
ROI mask image or volume, specified as a 2-D numeric matrix or medicalImage object for a medical image, or a 3-D numeric array or medicalVolume object for a medical volume. The data type of
roi must be the same as the data type of data.
If the ROI mask image or volume has multiple ROIs, you must label each ROI with a
different value to compute radiomics features separately for each ROI. This input
argument sets the ROILabel property of the radiomics
object.
Properties
This property is read-only.
Medical image or volume to prepare for radiomics, represented as a medicalImage
object or a medicalVolume object.
This property is read-only.
ROI mask image or volume, represented as a medicalImage
object or a medicalVolume object. The data type of ROILabel is the
same as the data type of Data. The
ROI mask image or volume can have multiple ROIs.
This property is read-only after object creation.
Medical volume and ROI mask volume resampling, specified as a numeric or logical
1 (true) or 0
(false). Specify Resample as
true to resample the medical volume and ROI mask volume to an
isotropic voxel spacing. When the Data and ROILabel properties of the radiomics object
are 3-D arrays or medicalVolume objects, the default value of Resample is
true.
Resampling is not applicable for 2-D data. When you specify the Data and ROILabel properties as 2-D matrices or medicalImage
objects, the default value is false, and you cannot change it.
Note
If you specify Resample as false, the
radiomics object ignores the values specified for the
ResampledVoxelSpacing, DataResampleMethod, and MaskResampleMethod properties.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
This property is read-only after object creation.
Medical image or volume resegmentation, specified as a numeric or logical
1 (true) or 0
(false). Specify Resegment as
true to resegment the medical image or volume.
Note
If you specify Resegment as false, the
radiomics object ignores the values specified for the
ResegmentationRange and ExcludeOutliers properties.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
This property is read-only after object creation.
Medical image or volume discretization, specified as a numeric or logical
1 (true) or 0
(false). Specify Discretize as
true to discretize the medical image or volume for computing
intensity histogram features and texture features.
Note
If you specify Discretize as false, the
radiomics object ignores the values specified for the
DiscreteBinSizeOrBinNumber and DiscreteMethod properties.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
This property is read-only after object creation.
Medical image or volume discretization for IVH features, specified as a numeric or
logical 1 (true) or 0
(false). Specify DiscretizeIVH as
true to discretize the medical image or volume for computing
intensity volume histogram (IVH) features.
Note
If you specify DiscretizeIVH as false, the
radiomics object ignores the values specified for the
DiscreteIVHBinSizeOrBinNumber and DiscreteIVHMethod properties.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
This property is read-only after object creation.
Resampled voxel spacing, specified a positive scalar. This value must be less than
half of the minimum spatial dimension of the medical volume. When the Data and ROILabel properties of the radiomics object
are 3-D arrays or medicalVolume objects, the default value of
ResampledVoxelSpacing is
ceil(min(data.VoxelSpacing)).
As resampling is not applicable for 2-D data, the radiomics
object ignores this value when you specify Data and ROILabel properties of the radiomics object
as 2-D matrices or medicalImage
objects.
Note
The radiomics object ignores this value if you specify
Resample as false.
Data Types: single | double
This property is read-only after object creation.
Medical volume resampling method, specified as "nearest",
"linear", "cubic", or
"spline". When the Data and ROILabel properties of the radiomics object
are 3-D arrays or medicalVolume objects, the default value of
DataResampleMethod is "linear".
As resampling is not applicable for 2-D data, the radiomics
object ignores this value when you specify Data and ROILabel properties of the radiomics object
as 2-D matrices or medicalImage
objects.
Note
The radiomics object ignores this value if you specify
Resample as false.
Data Types: char | string
This property is read-only after object creation.
ROI mask volume resampling method, specified as "nearest",
"linear", "cubic", or
"spline". When the Data and ROILabel properties of the radiomics object
are 3-D arrays or medicalVolume objects, the default value of
MaskResampleMethod is "linear".
As resampling is not applicable for 2-D data, the radiomics
object ignores this value when you specify Data and ROILabel properties of the radiomics object
as 2-D matrices or medicalImage
objects.
Note
The radiomics object ignores this value if you specify
Resample as false.
Data Types: char | string
This property is read-only after object creation.
Resegmentation range, specified as a two-element numeric vector. The object removes pixels or voxels with intensities outside the specified range. By default, the resegmentation range value is empty, and the object does not perform resegmentation.
Note
The radiomics object ignores this value if you specify
Resegment as false.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
This property is read-only after object creation.
Outlier exclusion, specified as a numeric or logical 1
(true) or 0 (false). Specify
ExcludeOutliers as true to compute the inlier
range of the medical image or volume and remove pixels or voxels with intensities
outside the inlier range.
Note
The radiomics object ignores this value if you specify
Resegment as false.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64 | logical
This property is read-only after object creation.
Bin size or bin number for discretization, specified as a positive scalar. The
functionality of this property differs based on the specified value of the
DiscreteMethod property.
"FixedBinSize"— The value ofDiscreteBinSizeOrNumberspecifies the bin size, and must be less than or equal to the maximum intensity value of the medical image or volume."FixedBinNumber"— The value ofDiscreteBinSizeOrNumberspecifies the bin number, and must be greater than 1.
By default, the object scales the range of the medical data scaled by a factor of 0.1 and rounds it, then performs discretization using the rounded range value as the bin size or bin number.
Note
The radiomics object ignores this value if you specify
Discretize as false.
Data Types: single | double
This property is read-only after object creation.
Discretization method, specified as "FixedBinSize" or
"FixedBinNumber".
Note
The radiomics object ignores this value if you specify
Discretize as false.
Data Types: char | string
This property is read-only after object creation.
Bin size or bin number for discretization for IVH features, specified as a positive
scalar. The functionality of this property differs based on the specified value of the
DiscreteIVHMethod property.
"FixedBinSize"— The value ofDiscreteIVHBinSizeOrNumberspecifies the bin size, and must be less than or equal to the maximum intensity value of the medical image or volume."FixedBinNumber"— The value ofDiscreteIVHBinSizeOrNumberspecifies the bin number, and must be greater than 1.
By default, the object scales the range of the medical data scaled by a factor of 0.1 and rounds it, then performs discretization using the rounded range value as the bin size or bin number.
Note
The radiomics object ignores this value if you specify
DiscretizeIVH as false.
Data Types: single | double | int8 | int16 | int32 | int64 | uint8 | uint16 | uint32 | uint64
This property is read-only after object creation.
Discretization method for IVH features, specified as
"FixedBinSize" or "FixedBinNumber".
Note
The radiomics object ignores this value if you specify
DiscretizeIVH as false.
Data Types: char | string
Object Functions
shapeFeatures | Radiomics shape features |
intensityFeatures | Radiomics intensity features |
textureFeatures | Radiomics texture features |
selectFeatures | Selected radiomics features |
Examples
Import a computed tomography (CT) image volume and the corresponding ROI mask volume from the IBSI validation data set [1][2][3] as medicalVolume objects.
unzip("CTImageMaskNIfTI.zip") data = medicalVolume("CT_image.nii.gz"); roi = medicalVolume("CT_mask.nii.gz");
Visualize a slice of the CT image volume and the corresponding ROI.
figure
imshowpair(data.Voxels(:,:,20),roi.Voxels(:,:,20),"montage")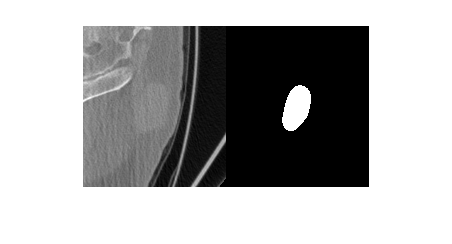
Create a radiomics object, using the CT image volume and ROI mask volume, with default preprocessing options.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalVolume]
ROILabel: [1×1 medicalVolume]
Resample: 1
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: 1
DataResampleMethod: 'linear'
MaskResampleMethod: 'linear'
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute the shape, intensity, and texture features of the ROI in the CT image volume.
S = shapeFeatures(R)
S=1×24 table
LabelID VolumeMesh3D VolumeVoxelCount3D SurfaceAreaMesh3D SurfaceVolumeRatio3D Compactness1_3D Compactness2_3D SphericalDisproportion3D Sphericity3D Asphericity3D CentreOfMassShift3D Maximum3dDiameter3D MajorAxisLength3D MinorAxisLength3D LeastAxisLength3D Elongation3D Flatness3D VolumeDensityAABB_3D AreaDensityAABB_3D VolumeDensityAEE_3D AreaDensityAEE_3D VolumeDensityConvexHull3D AreaDensityConvexHull3D IntegratedIntensity3D
_______ ____________ __________________ _________________ ____________________ _______________ _______________ ________________________ ____________ _____________ ___________________ ___________________ _________________ _________________ _________________ ____________ __________ ____________________ __________________ ___________________ _________________ _________________________ _______________________ _____________________
"1" 52923 52975 8425 0.15919 0.038611 0.5297 1.2359 0.80911 0.23592 0.8888 57.359 50.405 44.1 33.518 0.8749 0.66497 0.46924 0.59248 1.3566 1.4135 0.89714 1.0994 3.1069e+06
I = intensityFeatures(R)
I=1×50 table
LabelID LocalIntensityPeak3D GlobalIntensityPeak3D MeanIntensity3D IntensityVariance3D IntensitySkewness3D IntensityKurtosis3D MedianIntensity3D MinimumIntensity3D TenthIntensityPercentile3D NinetiethIntensityPercentile3D MaximumIntensity3D IntensityInterquartileRange3D IntensityRange3D MeanAbsoluteDeviation3D RobustMeanAbsoluteDeviation3D MedianAbsoluteDeviation3D CoefficientOfVariation3D QuartileCoefficientOfDispersion3D IntensityEnergy3D RootMeanSquare3D MeanDiscretisedIntensity3D DiscretisedIntensityVariance3D DiscretisedIntensitySkewness3D DiscretisedIntensityKurtosis3D MedianDiscretisedIntensity3D MinimumDiscretisedIntensity3D TenthDiscretisedIntensityPercentile3D NinetiethDiscretisedIntensityPercentile3D MaximumDiscretisedIntensity3D IntensityHistogramMode3D DiscretisedIntensityInterquartileRange3D DiscretisedIntensityRange3D IntensityHistogramMeanAbsoluteDeviation3D IntensityHistogramRobustMeanAbsoluteDeviation3D IntensityHistogramMedianAbsoluteDeviation3D IntensityHistogramCoeffcientOfVariation3D IntensityHistogramQuartileCoeffcientOfDispersion3D DiscretisedIntensityEntropy3D DiscretisedIntensityUniformity3D MaximumHistogramGradient3D MaximumHistogramGradientIntensity3D MinimumHistogramGradient3D MinimumHistogramGradientIntensity3D TenPercentVolumeFraction3D NinetyPercentVolumeFraction3D TenPercentIntensityFraction3D NinetyPercentIntensityFraction3D VolumeFractionDifference3D IntensityFractionDifference3D
_______ ____________________ _____________________ _______________ ___________________ ___________________ ___________________ _________________ __________________ __________________________ ______________________________ __________________ _____________________________ ________________ _______________________ _____________________________ _________________________ ________________________ _________________________________ _________________ ________________ __________________________ ______________________________ ______________________________ ______________________________ ____________________________ _____________________________ _____________________________________ _________________________________________ _____________________________ ________________________ ________________________________________ ___________________________ _________________________________________ _______________________________________________ ___________________________________________ _________________________________________ __________________________________________________ _____________________________ ________________________________ __________________________ ___________________________________ __________________________ ___________________________________ __________________________ _____________________________ _____________________________ ________________________________ __________________________ _____________________________
"1" 76.235 77.672 58.706 855.73 -0.11994 -0.04034 59 -31 21 96 148 39 179 23.302 16.403 23.294 0.4983 0.32773 2.2674e+08 65.588 9.5216 8.6378 -0.11707 -0.057331 10 1 6 13 18 10 4 17 2.3549 1.7176 2.3425 0.30867 0.2 3.5962 0.096231 1444 7 -1517.5 13 0.99048 0.0056728 711 292 0.9848 419
T = textureFeatures(R)
T=1×137 table
LabelID JointMaximumAveraged3D JointAverageAveraged3D JointVarianceAveraged3D JointEntropyAveraged3D DifferenceAverageAveraged3D DifferenceVarianceAveraged3D DifferenceEntropyAveraged3D SumAverageAveraged3D SumVarianceAveraged3D SumEntropyAveraged3D AngularSecondMomentAveraged3D ContrastAveraged3D DissimilarityAveraged3D InverseDifferenceAveraged3D NormalisedInverseDifferenceAveraged3D InverseDifferenceMomentAveraged3D NormalisedInverseDifferenceMomentAveraged3D InverseVarianceAveraged3D CorrelationAveraged3D AutoCorrelationAveraged3D ClusterTendencyAveraged3D ClusterShadeAveraged3D ClusterProminenceAveraged3D InformationCorrelation1Averaged3D InformationCorrelation2Averaged3D JointMaximumMerged3D JointAverageMerged3D JointVarianceMerged3D JointEntropyMerged3D DifferenceAverageMerged3D DifferenceVarianceMerged3D DifferenceEntropyMerged3D SumAverageMerged3D SumVarianceMerged3D SumEntropyMerged3D AngularSecondMomentMerged3D ContrastMerged3D DissimilarityMerged3D InverseDifferenceMerged3D NormalisedInverseDifferenceMerged3D InverseDifferenceMomentMerged3D NormalisedInverseDifferenceMomentMerged3D InverseVarianceMerged3D CorrelationMerged3D AutoCorrelationMerged3D ClusterTendencyMerged3D ClusterShadeMerged3D ClusterProminenceMerged3D InformationCorrelation1Merged3D InformationCorrelation2Merged3D ShortRunsEmphasisAveraged3D LongRunsEmphasisAveraged3D LowGrayLevelRunEmphasisAveraged3D HighGrayLevelRunEmphasisAveraged3D ShortRunLowGrayLevelEmphasisAveraged3D ShortRunHighGrayLevelEmphasisAveraged3D LongRunLowGrayLevelEmphasisAveraged3D LongRunHighGrayLevelEmphasisAveraged3D GrayLevelNonUniformityAveraged3D NormalisedGrayLevelNonUniformityAveraged3D RunLengthNonUniformityAveraged3D NormalisedRunLengthNonUniformityAveraged3D RunPercentageAveraged3D GrayLevelVarianceAveraged3D RunLengthVarianceAveraged3D RunEntropyAveraged3D ShortRunsEmphasisMerged3D LongRunsEmphasisMerged3D LowGrayLevelRunEmphasisMerged3D HighGrayLevelRunEmphasisMerged3D ShortRunLowGrayLevelEmphasisMerged3D ShortRunHighGrayLevelEmphasisMerged3D LongRunLowGrayLevelEmphasisMerged3D LongRunHighGrayLevelEmphasisMerged3D GrayLevelNonUniformityMerged3D NormalisedGrayLevelNonUniformityMerged3D RunLengthNonUniformityMerged3D NormalisedRunLengthNonUniformityMerged3D RunPercentageMerged3D GrayLevelVarianceMerged3D RunLengthVarianceMerged3D RunEntropyMerged3D SmallZoneEmphasis3D LargeZoneEmphasis3D LowGrayLevelZoneEmphasis3D HighGrayLevelZoneEmphasis3D SmallZoneLowGrayLevelEmphasis3D SmallZoneHighGrayLevelEmphasis3D LargeZoneLowGrayLevelEmphasis3D LargeZoneHighGrayLevelEmphasis3D GrayLevelNonUniformity3D NormalisedGrayLevelNonUniformity3D ZoneSizeNonUniformity3D NormalisedZoneSizeNonUniformity3D ZonePercentage3D GrayLevelVariance3D ZoneSizeVariance3D ZoneSizeEntropy3D SmallDistanceEmphasis3D LargeDistanceEmphasis3D LowGrayLevelZoneDistanceEmphasis3D HighGrayLevelZoneDistanceEmphasis3D SmallDistanceLowGrayLevelEmphasis3D SmallDistanceHighGrayLevelEmphasis3D LargeDistanceLowGrayLevelEmphasis3D LargeDistanceHighGrayLevelEmphasis3D GrayLevelDistanceNonUniformity3D NormalisedGrayLevelDistanceNonUniformity3D ZoneDistanceNonUniformity3D NormalisedZoneDistanceNonUniformity3D ZoneDistancePercentage3D GrayLevelDistanceVariance3D ZoneDistanceVariance3D ZoneDistanceEntropy3D Coarseness3D Contrast3D Busyness3D Complexity3D Strength3D LowDependenceEmphasis3D HighDependenceEmphasis3D LowGrayLevelCountEmphasis3D HighGrayLevelCountEmphasis3D LowDependenceLowGrayLevelEmphasis3D LowDependenceHighGrayLevelEmphasis3D HighDependenceLowGrayLevelEmphasis3D HighDependenceHighGrayLevelEmphasi3D GrayLevelDependenceNonUniformity3D NormalisedGrayLevelDependenceNonUniformity3D DependenceCountNonUniformity3D NormalisedDependenceCountNonUniformity3D DependenceCountPercentage3D GrayLevelDependenceVariance3D DependenceCountVariance3D DependenceCountEntropy3D DependenceCountEnergy3D
_______ ______________________ ______________________ _______________________ ______________________ ___________________________ ____________________________ ___________________________ ____________________ _____________________ ____________________ _____________________________ __________________ _______________________ ___________________________ _____________________________________ _________________________________ ___________________________________________ _________________________ _____________________ _________________________ _________________________ ______________________ ___________________________ _________________________________ _________________________________ ____________________ ____________________ _____________________ ____________________ _________________________ __________________________ _________________________ __________________ ___________________ __________________ ___________________________ ________________ _____________________ _________________________ ___________________________________ _______________________________ _________________________________________ _______________________ ___________________ _______________________ _______________________ ____________________ _________________________ _______________________________ _______________________________ ___________________________ __________________________ _________________________________ __________________________________ ______________________________________ _______________________________________ _____________________________________ ______________________________________ ________________________________ __________________________________________ ________________________________ __________________________________________ _______________________ ___________________________ ___________________________ ____________________ _________________________ ________________________ _______________________________ ________________________________ ____________________________________ _____________________________________ ___________________________________ ____________________________________ ______________________________ ________________________________________ ______________________________ ________________________________________ _____________________ _________________________ _________________________ __________________ ___________________ ___________________ __________________________ ___________________________ _______________________________ ________________________________ _______________________________ ________________________________ ________________________ __________________________________ _______________________ _________________________________ ________________ ___________________ __________________ _________________ _______________________ _______________________ __________________________________ ___________________________________ ___________________________________ ____________________________________ ___________________________________ ____________________________________ ________________________________ __________________________________________ ___________________________ _____________________________________ ________________________ ___________________________ ______________________ _____________________ ____________ __________ __________ ____________ __________ _______________________ ________________________ ___________________________ ____________________________ ___________________________________ ____________________________________ ____________________________________ ____________________________________ __________________________________ ____________________________________________ ______________________________ ________________________________________ ___________________________ _____________________________ _________________________ ________________________ _______________________
"1" 0.023811 9.6016 8.3563 6.8904 2.3407 3.5474 2.7164 19.203 24.082 4.3302 0.011903 9.3434 2.3407 0.42572 0.89254 0.34697 0.97367 0.34478 0.44066 95.876 24.082 -14.219 1766.6 -0.071978 0.51232 0.02387 9.6014 8.357 6.9797 2.3355 3.8557 2.796 19.203 24.118 4.3401 0.011132 9.3101 2.3355 0.42635 0.89275 0.34775 0.97376 0.34535 0.44298 95.889 24.118 -14.595 1772.3 -0.047062 0.53447 0.8948 1.6184 0.022051 99.343 0.020252 88.924 0.032272 160.51 4252.9 0.093867 34781 0.76314 0.85952 8.9998 0.24022 4.2908 0.89739 1.5847 0.022046 99.351 0.020307 89.195 0.031637 157.17 55285 0.093871 4.5075e+05 0.76535 0.85952 8.9979 0.23116 4.2962 0.54053 21751 0.044971 101.61 0.026927 55.289 237.38 2.1404e+06 398.92 0.065861 1677.4 0.27694 0.11492 18.452 21676 6.6843 0.37615 38.684 0.044971 101.61 0.023829 29.867 1.1708 4444.5 398.92 0.065861 901.04 0.14876 0.11492 18.452 16.962 7.3218 0.00015653 0.088117 36.374 194.83 0.015559 0.12867 26.96 0.021291 99.298 0.0046226 12.944 0.422 2663.2 5072.2 0.096231 6725.7 0.1276 1 8.6377 5.3148 6.6704 0.012506
Compute selected radiomics features for a 2-D slice of the CT image volume.
dataSlice = data.Voxels(:,:,20); roiSlice = roi.Voxels(:,:,20); R2D = radiomics(dataSlice,roiSlice)
R2D =
radiomics with properties:
Data: [1×1 medicalImage]
ROILabel: [1×1 medicalImage]
Resample: 0
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: []
DataResampleMethod: []
MaskResampleMethod: []
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
F = selectFeatures(R2D,["AreaDensityConvexHull2D","MaximumHistogramGradient2D","Complexity2D"])
F=1×4 table
LabelID AreaDensityConvexHull2D MaximumHistogramGradient2D Complexity2D
_______ _______________________ __________________________ ____________
"1" 0.97913 15 882.61
[1] Vallières, Martin, Carolyn R. Freeman, Sonia R. Skamene, and Issam El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” The Cancer Imaging Archive, 2015. https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS.
[2] Vallières, M, C R Freeman, S R Skamene, and I El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” Physics in Medicine and Biology 60, no. 14 (July 7, 2015): 5471–96. https://doi.org/10.1088/0031-9155/60/14/5471.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Import a magnetic resonance (MR) image volume from the IBSI validation data set [1][2][3] as a medicalVolume object.
unzip("MRImageMaskDICOM.zip") data = medicalVolume("MR_image");
Read the corresponding DICOM metadata from the DICOM-RT structure set file. Extract the ROI data from the structure set and ROI contour modules of the DICOM metadata.
info = dicominfo("MR_mask/RS.dcm");
contours = dicomContours(info);
contourIndex = 1;Get the spatial details of the DICOM volume. Create an ROI mask volume as a medicalVolume object from the spatial details and ROI contour.
[~,spatial] = dicomreadVolume("MR_image");
roi = createMask(contours,contourIndex,spatial);
roi = permute(roi,[2 1 3]);
roi = medicalVolume(uint8(roi),data.VolumeGeometry);Visualize a slice of the MR image volume and the corresponding ROI.
figure
imshowpair(data.Voxels(:,:,20),roi.Voxels(:,:,20),"montage")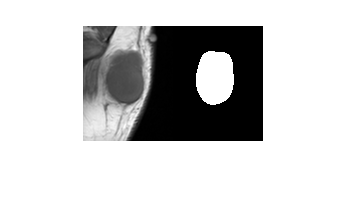
Create a radiomics object, using the MR image volume and ROI mask volume, with default preprocessing options.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalVolume]
ROILabel: [1×1 medicalVolume]
Resample: 1
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: 1
DataResampleMethod: 'linear'
MaskResampleMethod: 'linear'
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute the shape, intensity, and texture features of the ROI in the MR image volume.
S = shapeFeatures(R)
S=1×24 table
LabelID VolumeMesh3D VolumeVoxelCount3D SurfaceAreaMesh3D SurfaceVolumeRatio3D Compactness1_3D Compactness2_3D SphericalDisproportion3D Sphericity3D Asphericity3D CentreOfMassShift3D Maximum3dDiameter3D MajorAxisLength3D MinorAxisLength3D LeastAxisLength3D Elongation3D Flatness3D VolumeDensityAABB_3D AreaDensityAABB_3D VolumeDensityAEE_3D AreaDensityAEE_3D VolumeDensityConvexHull3D AreaDensityConvexHull3D IntegratedIntensity3D
_______ ____________ __________________ _________________ ____________________ _______________ _______________ ________________________ ____________ _____________ ___________________ ___________________ _________________ _________________ _________________ ____________ __________ ____________________ __________________ ___________________ _________________ _________________________ _______________________ _____________________
"1" 52977 53029 8267 0.15605 0.039764 0.5618 1.2119 0.82514 0.21192 0.58271 57.044 50.077 44.011 33.814 0.87885 0.67523 0.5173 0.62186 1.3577 1.3919 0.91396 1.092 1.9964e+07
I = intensityFeatures(R)
I=1×50 table
LabelID LocalIntensityPeak3D GlobalIntensityPeak3D MeanIntensity3D IntensityVariance3D IntensitySkewness3D IntensityKurtosis3D MedianIntensity3D MinimumIntensity3D TenthIntensityPercentile3D NinetiethIntensityPercentile3D MaximumIntensity3D IntensityInterquartileRange3D IntensityRange3D MeanAbsoluteDeviation3D RobustMeanAbsoluteDeviation3D MedianAbsoluteDeviation3D CoefficientOfVariation3D QuartileCoefficientOfDispersion3D IntensityEnergy3D RootMeanSquare3D MeanDiscretisedIntensity3D DiscretisedIntensityVariance3D DiscretisedIntensitySkewness3D DiscretisedIntensityKurtosis3D MedianDiscretisedIntensity3D MinimumDiscretisedIntensity3D TenthDiscretisedIntensityPercentile3D NinetiethDiscretisedIntensityPercentile3D MaximumDiscretisedIntensity3D IntensityHistogramMode3D DiscretisedIntensityInterquartileRange3D DiscretisedIntensityRange3D IntensityHistogramMeanAbsoluteDeviation3D IntensityHistogramRobustMeanAbsoluteDeviation3D IntensityHistogramMedianAbsoluteDeviation3D IntensityHistogramCoeffcientOfVariation3D IntensityHistogramQuartileCoeffcientOfDispersion3D DiscretisedIntensityEntropy3D DiscretisedIntensityUniformity3D MaximumHistogramGradient3D MaximumHistogramGradientIntensity3D MinimumHistogramGradient3D MinimumHistogramGradientIntensity3D TenPercentVolumeFraction3D NinetyPercentVolumeFraction3D TenPercentIntensityFraction3D NinetyPercentIntensityFraction3D VolumeFractionDifference3D IntensityFractionDifference3D
_______ ____________________ _____________________ _______________ ___________________ ___________________ ___________________ _________________ __________________ __________________________ ______________________________ __________________ _____________________________ ________________ _______________________ _____________________________ _________________________ ________________________ _________________________________ _________________ ________________ __________________________ ______________________________ ______________________________ ______________________________ ____________________________ _____________________________ _____________________________________ _________________________________________ _____________________________ ________________________ ________________________________________ ___________________________ _________________________________________ _______________________________________________ ___________________________________________ _________________________________________ __________________________________________________ _____________________________ ________________________________ __________________________ ___________________________________ __________________________ ___________________________________ __________________________ _____________________________ _____________________________ ________________________________ __________________________ _____________________________
"1" 716.07 716.07 376.84 5820.4 1.6378 2.2078 351 265 311 495 662 72 397 56.834 34.043 51.62 0.20245 0.099448 7.6772e+09 384.49 11.766 59.148 1.6361 2.1973 9 1 5 24 40 7 7 39 5.7308 3.5845 5.1877 0.65364 0.33333 4.4234 0.064718 2057.5 4 -966.5 10 0.95596 0.01398 581 117 0.94198 464
T = textureFeatures(R)
T=1×137 table
LabelID JointMaximumAveraged3D JointAverageAveraged3D JointVarianceAveraged3D JointEntropyAveraged3D DifferenceAverageAveraged3D DifferenceVarianceAveraged3D DifferenceEntropyAveraged3D SumAverageAveraged3D SumVarianceAveraged3D SumEntropyAveraged3D AngularSecondMomentAveraged3D ContrastAveraged3D DissimilarityAveraged3D InverseDifferenceAveraged3D NormalisedInverseDifferenceAveraged3D InverseDifferenceMomentAveraged3D NormalisedInverseDifferenceMomentAveraged3D InverseVarianceAveraged3D CorrelationAveraged3D AutoCorrelationAveraged3D ClusterTendencyAveraged3D ClusterShadeAveraged3D ClusterProminenceAveraged3D InformationCorrelation1Averaged3D InformationCorrelation2Averaged3D JointMaximumMerged3D JointAverageMerged3D JointVarianceMerged3D JointEntropyMerged3D DifferenceAverageMerged3D DifferenceVarianceMerged3D DifferenceEntropyMerged3D SumAverageMerged3D SumVarianceMerged3D SumEntropyMerged3D AngularSecondMomentMerged3D ContrastMerged3D DissimilarityMerged3D InverseDifferenceMerged3D NormalisedInverseDifferenceMerged3D InverseDifferenceMomentMerged3D NormalisedInverseDifferenceMomentMerged3D InverseVarianceMerged3D CorrelationMerged3D AutoCorrelationMerged3D ClusterTendencyMerged3D ClusterShadeMerged3D ClusterProminenceMerged3D InformationCorrelation1Merged3D InformationCorrelation2Merged3D ShortRunsEmphasisAveraged3D LongRunsEmphasisAveraged3D LowGrayLevelRunEmphasisAveraged3D HighGrayLevelRunEmphasisAveraged3D ShortRunLowGrayLevelEmphasisAveraged3D ShortRunHighGrayLevelEmphasisAveraged3D LongRunLowGrayLevelEmphasisAveraged3D LongRunHighGrayLevelEmphasisAveraged3D GrayLevelNonUniformityAveraged3D NormalisedGrayLevelNonUniformityAveraged3D RunLengthNonUniformityAveraged3D NormalisedRunLengthNonUniformityAveraged3D RunPercentageAveraged3D GrayLevelVarianceAveraged3D RunLengthVarianceAveraged3D RunEntropyAveraged3D ShortRunsEmphasisMerged3D LongRunsEmphasisMerged3D LowGrayLevelRunEmphasisMerged3D HighGrayLevelRunEmphasisMerged3D ShortRunLowGrayLevelEmphasisMerged3D ShortRunHighGrayLevelEmphasisMerged3D LongRunLowGrayLevelEmphasisMerged3D LongRunHighGrayLevelEmphasisMerged3D GrayLevelNonUniformityMerged3D NormalisedGrayLevelNonUniformityMerged3D RunLengthNonUniformityMerged3D NormalisedRunLengthNonUniformityMerged3D RunPercentageMerged3D GrayLevelVarianceMerged3D RunLengthVarianceMerged3D RunEntropyMerged3D SmallZoneEmphasis3D LargeZoneEmphasis3D LowGrayLevelZoneEmphasis3D HighGrayLevelZoneEmphasis3D SmallZoneLowGrayLevelEmphasis3D SmallZoneHighGrayLevelEmphasis3D LargeZoneLowGrayLevelEmphasis3D LargeZoneHighGrayLevelEmphasis3D GrayLevelNonUniformity3D NormalisedGrayLevelNonUniformity3D ZoneSizeNonUniformity3D NormalisedZoneSizeNonUniformity3D ZonePercentage3D GrayLevelVariance3D ZoneSizeVariance3D ZoneSizeEntropy3D SmallDistanceEmphasis3D LargeDistanceEmphasis3D LowGrayLevelZoneDistanceEmphasis3D HighGrayLevelZoneDistanceEmphasis3D SmallDistanceLowGrayLevelEmphasis3D SmallDistanceHighGrayLevelEmphasis3D LargeDistanceLowGrayLevelEmphasis3D LargeDistanceHighGrayLevelEmphasis3D GrayLevelDistanceNonUniformity3D NormalisedGrayLevelDistanceNonUniformity3D ZoneDistanceNonUniformity3D NormalisedZoneDistanceNonUniformity3D ZoneDistancePercentage3D GrayLevelDistanceVariance3D ZoneDistanceVariance3D ZoneDistanceEntropy3D Coarseness3D Contrast3D Busyness3D Complexity3D Strength3D LowDependenceEmphasis3D HighDependenceEmphasis3D LowGrayLevelCountEmphasis3D HighGrayLevelCountEmphasis3D LowDependenceLowGrayLevelEmphasis3D LowDependenceHighGrayLevelEmphasis3D HighDependenceLowGrayLevelEmphasis3D HighDependenceHighGrayLevelEmphasi3D GrayLevelDependenceNonUniformity3D NormalisedGrayLevelDependenceNonUniformity3D DependenceCountNonUniformity3D NormalisedDependenceCountNonUniformity3D DependenceCountPercentage3D GrayLevelDependenceVariance3D DependenceCountVariance3D DependenceCountEntropy3D DependenceCountEnergy3D
_______ ______________________ ______________________ _______________________ ______________________ ___________________________ ____________________________ ___________________________ ____________________ _____________________ ____________________ _____________________________ __________________ _______________________ ___________________________ _____________________________________ _________________________________ ___________________________________________ _________________________ _____________________ _________________________ _________________________ ______________________ ___________________________ _________________________________ _________________________________ ____________________ ____________________ _____________________ ____________________ _________________________ __________________________ _________________________ __________________ ___________________ __________________ ___________________________ ________________ _____________________ _________________________ ___________________________________ _______________________________ _________________________________________ _______________________ ___________________ _______________________ _______________________ ____________________ _________________________ _______________________________ _______________________________ ___________________________ __________________________ _________________________________ __________________________________ ______________________________________ _______________________________________ _____________________________________ ______________________________________ ________________________________ __________________________________________ ________________________________ __________________________________________ _______________________ ___________________________ ___________________________ ____________________ _________________________ ________________________ _______________________________ ________________________________ ____________________________________ _____________________________________ ___________________________________ ____________________________________ ______________________________ ________________________________________ ______________________________ ________________________________________ _____________________ _________________________ _________________________ __________________ ___________________ ___________________ __________________________ ___________________________ _______________________________ ________________________________ _______________________________ ________________________________ ________________________ __________________________________ _______________________ _________________________________ ________________ ___________________ __________________ _________________ _______________________ _______________________ __________________________________ ___________________________________ ___________________________________ ____________________________________ ___________________________________ ____________________________________ ________________________________ __________________________________________ ___________________________ _____________________________________ ________________________ ___________________________ ______________________ _____________________ ____________ __________ __________ ____________ __________ _______________________ ________________________ ___________________________ ____________________________ ___________________________________ ____________________________________ ____________________________________ ____________________________________ __________________________________ ____________________________________________ ______________________________ ________________________________________ ___________________________ _____________________________ _________________________ ________________________ _______________________
"1" 0.055201 11.152 49.523 7.1139 1.7744 6.5868 2.5284 22.304 188.25 5.279 0.020007 9.8416 1.7744 0.58486 0.96071 0.54425 0.99421 0.37181 0.89929 168.99 188.25 4341 1.9593e+05 -0.35169 0.97251 0.054921 11.154 49.561 7.1632 1.7706 6.6692 2.5465 22.307 188.44 5.2854 0.019688 9.8041 1.7706 0.58533 0.96079 0.54477 0.99423 0.3717 0.90109 169.06 188.44 4353.5 1.9663e+05 -0.34081 0.97328 0.79186 3.639 0.014285 255.46 0.0097224 235.58 0.077719 443.56 1736.7 0.049595 20497 0.58478 0.672 70.681 1.3817 5.8533 0.79358 3.5532 0.014342 254.45 0.0098114 234.83 0.07564 436.57 22493 0.049578 2.6594e+05 0.58617 0.672 70.759 1.3388 5.8673 0.62345 23996 0.0044919 582.9 0.0023151 382.95 514.26 1.4324e+06 199.65 0.032686 2210.5 0.36191 0.11761 71.698 23924 7.1635 0.58649 5.8012 0.0044919 582.9 0.0019566 378.98 0.057854 2508.2 199.65 0.032686 2057.9 0.33692 0.11761 71.698 1.904 6.9196 0.00051829 0.070477 5.531 610.45 0.44531 0.11335 135.06 0.017226 197.59 0.00062986 59.623 3.3416 8548.8 3361 0.064718 2707.1 0.052127 1 59.146 44.279 8.2752 0.0038749
Compute selected radiomics features for a 2-D slice of the MR image volume.
dataSlice = data.Voxels(:,:,20); roiSlice = roi.Voxels(:,:,20); R2D = radiomics(dataSlice,roiSlice)
R2D =
radiomics with properties:
Data: [1×1 medicalImage]
ROILabel: [1×1 medicalImage]
Resample: 0
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: []
DataResampleMethod: []
MaskResampleMethod: []
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
F = selectFeatures(R2D,["SphericalDisproportion2D","LocalIntensityPeak2D","IntensityVariance2D","DifferenceEntropyAveraged2D"])
F=1×5 table
LabelID SphericalDisproportion2D LocalIntensityPeak2D IntensityVariance2D DifferenceEntropyAveraged2D
_______ ________________________ ____________________ ___________________ ___________________________
"1" 5.0986 614.26 2140.8 2.1156
[1] Vallières, Martin, Carolyn R. Freeman, Sonia R. Skamene, and Issam El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” The Cancer Imaging Archive, 2015. https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS.
[2] Vallières, M, C R Freeman, S R Skamene, and I El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” Physics in Medicine and Biology 60, no. 14 (July 7, 2015): 5471–96. https://doi.org/10.1088/0031-9155/60/14/5471.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Version History
Introduced in R2023bCreate a radiomics object for 2-D medical data, specified as a 2-D
matrix or medicalImage object. Additionally, you can now create a
radiomics object for 3-D medical data by specifying a 3-D array.
Compute selected radiomics features using the selectFeatures object
function.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)