textureFeatures
Description
Examples
Load an X-ray image into the workspace as a medicalImage object. Visualize the image.
data = medicalImage("forearmXrayImage1.dcm");
I = data.Pixels;
figure
imshow(I,[])Draw two regions of interest (ROI) in the X-ray image. Create masks from the ROIs.
roi1 = drawassisted(Color="g"); roi2 = drawassisted(Color="r");

mask1 = createMask(roi1,I); mask2 = createMask(roi2,I);
Create an ROI label matrix, using different labels for the two ROIs. Create a medicalImage object of the ROI label data.
mask = zeros(size(I));
mask(mask1) = 1;
mask(mask2) = 2;
info = dicominfo("forearmXrayImage1.dcm");
roi = medicalImage(mask,info);Create a radiomics object from the X-ray image data and ROI label data.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalImage]
ROILabel: [1×1 medicalImage]
Resample: 0
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: []
DataResampleMethod: []
MaskResampleMethod: []
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute texture features for both ROIs.
T = textureFeatures(R)
T=2×137 table
LabelID JointMaximumAveraged2D JointAverageAveraged2D JointVarianceAveraged2D JointEntropyAveraged2D DifferenceAverageAveraged2D DifferenceVarianceAveraged2D DifferenceEntropyAveraged2D SumAverageAveraged2D SumVarianceAveraged2D SumEntropyAveraged2D AngularSecondMomentAveraged2D ContrastAveraged2D DissimilarityAveraged2D InverseDifferenceAveraged2D NormalisedInverseDifferenceAveraged2D InverseDifferenceMomentAveraged2D NormalisedInverseDifferenceMomentAveraged2D InverseVarianceAveraged2D CorrelationAveraged2D AutoCorrelationAveraged2D ClusterTendencyAveraged2D ClusterShadeAveraged2D ClusterProminenceAveraged2D InformationCorrelation1Averaged2D InformationCorrelation2Averaged2D JointMaximumSliceMerged2D JointAverageSliceMerged2D JointVarianceSliceMerged2D JointEntropySliceMerged2D DifferenceAverageSliceMerged2D DifferenceVarianceSliceMerged2D DifferenceEntropySliceMerged2D SumAverageSliceMerged2D SumVarianceSliceMerged2D SumEntropySliceMerged2D AngularSecondMomentSliceMerged2D ContrastSliceMerged2D DissimilaritySliceMerged2D InverseDifferenceSliceMerged2D NormalisedInverseDifferenceSliceMerged2D InverseDifferenceMomentSliceMerged2D NormalisedInverseDifferenceMomentSliceMerged2D InverseVarianceSliceMerged2D CorrelationSliceMerged2D AutoCorrelationSliceMerged2D ClusterTendencySliceMerged2D ClusterShadeSliceMerged2D ClusterProminenceSliceMerged2D InformationCorrelation1SliceMerged2D InformationCorrelation2SliceMerged2D ShortRunsEmphasisAveraged2D LongRunsEmphasisAveraged2D LowGrayLevelRunEmphasisAveraged2D HighGrayLevelRunEmphasisAveraged2D ShortRunLowGrayLevelEmphasisAveraged2D ShortRunHighGrayLevelEmphasisAveraged2D LongRunLowGrayLevelEmphasisAveraged2D LongRunHighGrayLevelEmphasisAveraged2D GrayLevelNonUniformityAveraged2D NormalisedGrayLevelNonUniformityAveraged2D RunLengthNonUniformityAveraged2D NormalisedRunLengthNonUniformityAveraged2D RunPercentageAveraged2D GrayLevelVarianceAveraged2D RunLengthVarianceAveraged2D RunEntropyAveraged2D ShortRunsEmphasisSliceMerged2D LongRunsEmphasisSliceMerged2D LowGrayLevelRunEmphasisSliceMerged2D HighGrayLevelRunEmphasisSliceMerged2D ShortRunLowGrayLevelEmphasisSliceMerged2D ShortRunHighGrayLevelEmphasisSliceMerged2D LongRunLowGrayLevelEmphasisSliceMerged2D LongRunHighGrayLevelEmphasisSliceMerged2D GrayLevelNonUniformitySliceMerged2D NormalisedGrayLevelNonUniformitySliceMerged2D RunLengthNonUniformitySliceMerged2D NormalisedRunLengthNonUniformitySliceMerged2D RunPercentageSliceMerged2D GrayLevelVarianceSliceMerged2D RunLengthVarianceSliceMerged2D RunEntropySliceMerged2D SmallZoneEmphasis2D LargeZoneEmphasis2D LowGrayLevelZoneEmphasis2D HighGrayLevelZoneEmphasis2D SmallZoneLowGrayLevelEmphasis2D SmallZoneHighGrayLevelEmphasis2D LargeZoneLowGrayLevelEmphasis2D LargeZoneHighGrayLevelEmphasis2D GrayLevelNonUniformity2D NormalisedGrayLevelNonUniformity2D ZoneSizeNonUniformity2D NormalisedZoneSizeNonUniformity2D ZonePercentage2D GrayLevelVariance2D ZoneSizeVariance2D ZoneSizeEntropy2D SmallDistanceEmphasis2D LargeDistanceEmphasis2D LowGrayLevelDistanceZoneEmphasis2D HighGrayLevelDistanceZoneEmphasis2D SmallDistanceLowGrayLevelEmphasis2D SmallDistanceHighGrayLevelEmphasis2D LargeDistanceLowGrayLevelEmphasis2D LargeDistanceHighGrayLevelEmphasis2D GrayLevelDistanceNonUniformity2D NormalisedGrayLevelDistanceNonUniformity2D ZoneDistanceNonUniformity2D NormalisedZoneDistanceNonUniformity2D ZoneDistancePercentage2D GrayLevelDistanceVariance2D ZoneDistanceVariance2D ZoneDistanceEntropy2D Coarseness2D Contrast2D Busyness2D Complexity2D Strength2D LowDependenceEmphasis2D HighDependenceEmphasis2D LowGrayLevelCountEmphasis2D HighGrayLevelCountEmphasis2D LowDependenceLowGrayLevelEmphasis2D LowDependenceHighGrayLevelEmphasis2D HighDependenceLowGrayLevelEmphasis2D HighDependenceHighGrayLevelEmphasis2D GrayLevelDependenceNonUniformity2D NormalisedGrayLevelDependenceNonUniformity2D DependenceCountNonUniformity2D NormalisedDependenceCountNonUniformity2D DependenceCountPercentage2D GrayLevelDependenceVariance2D DependenceCountVariance2D DependenceCountEntropy2D DependenceCountEnergy2D
_______ ______________________ ______________________ _______________________ ______________________ ___________________________ ____________________________ ___________________________ ____________________ _____________________ ____________________ _____________________________ __________________ _______________________ ___________________________ _____________________________________ _________________________________ ___________________________________________ _________________________ _____________________ _________________________ _________________________ ______________________ ___________________________ _________________________________ _________________________________ _________________________ _________________________ __________________________ _________________________ ______________________________ _______________________________ ______________________________ _______________________ ________________________ _______________________ ________________________________ _____________________ __________________________ ______________________________ ________________________________________ ____________________________________ ______________________________________________ ____________________________ ________________________ ____________________________ ____________________________ _________________________ ______________________________ ____________________________________ ____________________________________ ___________________________ __________________________ _________________________________ __________________________________ ______________________________________ _______________________________________ _____________________________________ ______________________________________ ________________________________ __________________________________________ ________________________________ __________________________________________ _______________________ ___________________________ ___________________________ ____________________ ______________________________ _____________________________ ____________________________________ _____________________________________ _________________________________________ __________________________________________ ________________________________________ _________________________________________ ___________________________________ _____________________________________________ ___________________________________ _____________________________________________ __________________________ ______________________________ ______________________________ _______________________ ___________________ ___________________ __________________________ ___________________________ _______________________________ ________________________________ _______________________________ ________________________________ ________________________ __________________________________ _______________________ _________________________________ ________________ ___________________ __________________ _________________ _______________________ _______________________ __________________________________ ___________________________________ ___________________________________ ____________________________________ ___________________________________ ____________________________________ ________________________________ __________________________________________ ___________________________ _____________________________________ ________________________ ___________________________ ______________________ _____________________ ____________ __________ __________ ____________ __________ _______________________ ________________________ ___________________________ ____________________________ ___________________________________ ____________________________________ ____________________________________ _____________________________________ __________________________________ ____________________________________________ ______________________________ ________________________________________ ___________________________ _____________________________ _________________________ ________________________ _______________________
"1" 0.0024655 57.173 675.82 11.068 8.6729 46.565 4.5233 114.35 2580.4 7.4309 0.00059926 122.85 8.6729 0.19708 0.94922 0.11252 0.9951 0.11632 0.90898 3883.2 2580.4 1.3078e+05 2.3618e+07 -0.29751 0.98931 0.001644 57.174 675.85 11.596 8.6687 47.58 4.548 114.35 2580.7 7.4877 0.00046409 122.73 8.6687 0.19713 0.94925 0.11255 0.9951 0.11635 0.90921 3883.3 2580.7 1.3082e+05 2.3623e+07 -0.21644 0.96957 0.97102 1.1229 0.00098018 4026.2 0.0009635 3914 0.0010494 4515.4 36.557 0.013495 2509.3 0.92621 0.96166 695.41 0.041421 6.7308 0.97106 1.1227 0.00098015 4026.1 0.00096349 3914.1 0.0010493 4514.8 146.12 0.013485 10037 0.92622 0.96166 695.39 0.041415 6.7564 0.89606 1.5878 0.0010299 4044.5 0.00097106 3647.6 0.0013676 6380.2 31.872 0.013203 1842 0.76303 0.85694 692.9 0.22607 7.1293 0.10493 148.22 0.0010299 4044.5 4.2891e-05 687.52 0.25647 3.5734e+05 31.872 0.013203 111.25 0.046084 0.85694 692.9 44.223 10.151 0.0057315 0.36515 0.026994 41509 53.247 0.80061 2.0302 0.00096438 4022.6 0.00085099 3251.6 0.0015501 8140.3 38.259 0.013582 1686.2 0.59857 1 696.64 0.32268 7.3893 0.0080629
"2" 0.0020617 95.454 1035.3 11.429 7.9161 41.032 4.4079 190.91 4036.8 7.7995 0.00046008 104.57 7.9161 0.21086 0.9534 0.12386 0.99582 0.12644 0.94946 10095 4036.8 -1.9066e+05 4.8383e+07 -0.32989 0.99441 0.001596 95.454 1035.4 11.899 7.9126 41.871 4.4312 190.91 4036.9 7.8439 0.00036457 104.48 7.9126 0.21092 0.95342 0.12391 0.99583 0.12649 0.94954 10095 4036.9 -1.9069e+05 4.8387e+07 -0.26141 0.98594 0.96734 1.1406 0.0025825 10034 0.0025562 9680.6 0.0026925 11556 37.312 0.010109 3385.9 0.91727 0.95671 1070 0.047944 7.0955 0.96737 1.1405 0.0025823 10034 0.002556 9680.9 0.0026925 11555 149.12 0.0101 13543 0.91728 0.95671 1070 0.047939 7.12 0.87894 1.6924 0.0028339 9925.6 0.0027386 8633.6 0.0034115 17474 31.832 0.0098705 2354 0.72991 0.83593 1076.5 0.26132 7.5598 0.088426 218.2 0.0028339 9925.6 0.0013534 674.8 0.05072 2.5181e+06 31.832 0.0098705 121.73 0.037744 0.83593 1076.5 66.086 10.766 0.0059013 0.39908 0.010539 44586 60.208 0.77294 2.1524 0.002505 10068 0.0023228 7628.5 0.0034117 22598 39.306 0.010188 2164.5 0.56105 1 1067.5 0.3399 7.8224 0.0056895
Import a computed tomography (CT) image volume and the corresponding ROI mask volume from the IBSI validation data set [1][2][3] as medicalVolume objects.
unzip("CTImageMaskNIfTI.zip") data = medicalVolume("CT_image.nii.gz"); roi = medicalVolume("CT_mask.nii.gz");
Visualize a slice of the CT image volume and the corresponding ROI.
figure
imshowpair(data.Voxels(:,:,20),roi.Voxels(:,:,20),"montage")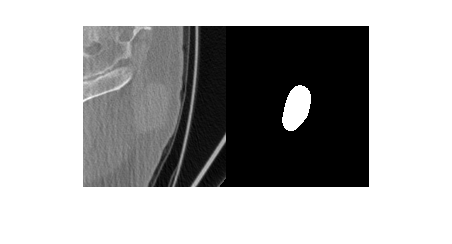
Create a radiomics object, using the CT image volume and ROI mask volume, with default preprocessing options.
R = radiomics(data,roi)
R =
radiomics with properties:
Data: [1×1 medicalVolume]
ROILabel: [1×1 medicalVolume]
Resample: 1
Resegment: 1
Discretize: 1
DiscretizeIVH: 1
ResampledVoxelSpacing: 1
DataResampleMethod: 'linear'
MaskResampleMethod: 'linear'
ResegmentationRange: []
ExcludeOutliers: 1
DiscreteBinSizeOrBinNumber: []
DiscreteMethod: 'FixedBinNumber'
DiscreteIVHBinSizeOrBinNumber: []
DiscreteIVHMethod: 'FixedBinNumber'
Compute the grey level co-occurrence matrix (GLCM) texture features of the ROI in the 2-D resampled CT image volume.
I = textureFeatures(R,Type=["GLCM","GLRLM"],SubType="2D")
I=1×83 table
LabelID JointMaximumAveraged2D JointAverageAveraged2D JointVarianceAveraged2D JointEntropyAveraged2D DifferenceAverageAveraged2D DifferenceVarianceAveraged2D DifferenceEntropyAveraged2D SumAverageAveraged2D SumVarianceAveraged2D SumEntropyAveraged2D AngularSecondMomentAveraged2D ContrastAveraged2D DissimilarityAveraged2D InverseDifferenceAveraged2D NormalisedInverseDifferenceAveraged2D InverseDifferenceMomentAveraged2D NormalisedInverseDifferenceMomentAveraged2D InverseVarianceAveraged2D CorrelationAveraged2D AutoCorrelationAveraged2D ClusterTendencyAveraged2D ClusterShadeAveraged2D ClusterProminenceAveraged2D InformationCorrelation1Averaged2D InformationCorrelation2Averaged2D JointMaximumSliceMerged2D JointAverageSliceMerged2D JointVarianceSliceMerged2D JointEntropySliceMerged2D DifferenceAverageSliceMerged2D DifferenceVarianceSliceMerged2D DifferenceEntropySliceMerged2D SumAverageSliceMerged2D SumVarianceSliceMerged2D SumEntropySliceMerged2D AngularSecondMomentSliceMerged2D ContrastSliceMerged2D DissimilaritySliceMerged2D InverseDifferenceSliceMerged2D NormalisedInverseDifferenceSliceMerged2D InverseDifferenceMomentSliceMerged2D NormalisedInverseDifferenceMomentSliceMerged2D InverseVarianceSliceMerged2D CorrelationSliceMerged2D AutoCorrelationSliceMerged2D ClusterTendencySliceMerged2D ClusterShadeSliceMerged2D ClusterProminenceSliceMerged2D InformationCorrelation1SliceMerged2D InformationCorrelation2SliceMerged2D ShortRunsEmphasisAveraged2D LongRunsEmphasisAveraged2D LowGrayLevelRunEmphasisAveraged2D HighGrayLevelRunEmphasisAveraged2D ShortRunLowGrayLevelEmphasisAveraged2D ShortRunHighGrayLevelEmphasisAveraged2D LongRunLowGrayLevelEmphasisAveraged2D LongRunHighGrayLevelEmphasisAveraged2D GrayLevelNonUniformityAveraged2D NormalisedGrayLevelNonUniformityAveraged2D RunLengthNonUniformityAveraged2D NormalisedRunLengthNonUniformityAveraged2D RunPercentageAveraged2D GrayLevelVarianceAveraged2D RunLengthVarianceAveraged2D RunEntropyAveraged2D ShortRunsEmphasisSliceMerged2D LongRunsEmphasisSliceMerged2D LowGrayLevelRunEmphasisSliceMerged2D HighGrayLevelRunEmphasisSliceMerged2D ShortRunLowGrayLevelEmphasisSliceMerged2D ShortRunHighGrayLevelEmphasisSliceMerged2D LongRunLowGrayLevelEmphasisSliceMerged2D LongRunHighGrayLevelEmphasisSliceMerged2D GrayLevelNonUniformitySliceMerged2D NormalisedGrayLevelNonUniformitySliceMerged2D RunLengthNonUniformitySliceMerged2D NormalisedRunLengthNonUniformitySliceMerged2D RunPercentageSliceMerged2D GrayLevelVarianceSliceMerged2D RunLengthVarianceSliceMerged2D RunEntropySliceMerged2D
_______ ______________________ ______________________ _______________________ ______________________ ___________________________ ____________________________ ___________________________ ____________________ _____________________ ____________________ _____________________________ __________________ _______________________ ___________________________ _____________________________________ _________________________________ ___________________________________________ _________________________ _____________________ _________________________ _________________________ ______________________ ___________________________ _________________________________ _________________________________ _________________________ _________________________ __________________________ _________________________ ______________________________ _______________________________ ______________________________ _______________________ ________________________ _______________________ ________________________________ _____________________ __________________________ ______________________________ ________________________________________ ____________________________________ ______________________________________________ ____________________________ ________________________ ____________________________ ____________________________ _________________________ ______________________________ ____________________________________ ____________________________________ ___________________________ __________________________ _________________________________ __________________________________ ______________________________________ _______________________________________ _____________________________________ ______________________________________ ________________________________ __________________________________________ ________________________________ __________________________________________ _______________________ ___________________________ ___________________________ ____________________ ______________________________ _____________________________ ____________________________________ _____________________________________ _________________________________________ __________________________________________ ________________________________________ _________________________________________ ___________________________________ _____________________________________________ ___________________________________ _____________________________________________ __________________________ ______________________________ ______________________________ _______________________
"1" 0.025292 10.263 9.9482 6.9415 2.6834 4.353 2.8719 20.525 27.92 4.3458 0.011452 11.872 2.6834 0.39071 0.8891 0.3042 0.97287 0.30871 0.39971 111.04 27.92 -17.682 2401.8 -0.10434 0.66674 0.021298 10.262 9.9491 7.1238 2.678 4.5887 2.9277 20.525 27.962 4.3981 0.0098741 11.835 2.678 0.39129 0.8893 0.3049 0.97295 0.30908 0.40201 111.06 27.962 -18.094 2408.5 -0.052692 0.55415 0.91083 1.4376 0.023078 119.32 0.021376 108.52 0.030767 173.59 75.51 0.087674 710.23 0.79414 0.896 12.076 0.15199 4.243 0.91189 1.4316 0.023109 119.34 0.021486 108.64 0.030465 172.87 301.76 0.087414 2836.1 0.79414 0.896 12.083 0.15192 4.2827
[1] Vallières, Martin, Carolyn R. Freeman, Sonia R. Skamene, and Issam El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” The Cancer Imaging Archive, 2015. https://doi.org/10.7937/K9/TCIA.2015.7GO2GSKS.
[2] Vallières, M, C R Freeman, S R Skamene, and I El Naqa. “A Radiomics Model from Joint FDG-PET and MRI Texture Features for the Prediction of Lung Metastases in Soft-Tissue Sarcomas of the Extremities.” Physics in Medicine and Biology 60, no. 14 (July 7, 2015): 5471–96. https://doi.org/10.1088/0031-9155/60/14/5471.
[3] Clark, Kenneth, Bruce Vendt, Kirk Smith, John Freymann, Justin Kirby, Paul Koppel, Stephen Moore, et al. “The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository.” Journal of Digital Imaging 26, no. 6 (December 2013): 1045–57. https://doi.org/10.1007/s10278-013-9622-7.
Input Arguments
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Example: textureFeatures(R,Type="GLCM",SubType="2D") computes the GLCM
texture features as the average over 2-D slices in the 2-D resampled data.
Category of texture features to compute, specified as one or more of these options.
"GLCM"— Grey level co-occurrence matrix"GLRLM"— Grey level run length matrix"GLSZM"— Grey level size zone matrix"GLDZM"— Grey level distance zone matrix"NGTDM"— Neighbourhood grey tone difference matrix"NGLDM"— Neighbourhood grey level dependence matrix"all"
If you specify "all", the function computes every
category of texture features. For more information on which specific texture features
each category includes, see IBSI Standard and Radiomics Function Feature Correspondences.
Data Types: char | string
Resampling from which to compute texture features, specified as one of these options.
"2D"— Computes features for each 2-D slice in the 2-D resampled data and averages them. This is the default option for 2-D data, when theDataandROILabelproperties of theradiomicsobjectRare 2-D matrices ormedicalImageobjects."2.5D"— Computes features after merging all 2-D slices in the 2-D resampled data."3D"— Computes features for the entire 3-D volume in the 3-D resampled data. This is the default option for 3-D data, when theDataandROILabelproperties of theradiomicsobjectRare 3-D arrays ormedicalVolumeobjects. 3-D resampling is not applicable for 2-D data."all"— Computes features for all applicable options.
For 3-D data, when you perform 3-D resampling of the volume, the function makes the voxel spacing along all three spatial dimensions isotropic. However, when you perform 2-D resampling of the volume, the function makes the voxel spacing along only the x- and y-dimensions isotropic, while retaining the voxel spacing of the input volume along the z-dimension.
Data Types: char | string
Output Arguments
Texture features, returned as a table. The first column in T is
LabelID. The subsequent columns are the texture features. Each row
of the table corresponds to an ROI. For more details on which texture features are
computed in each Type and SubType, see IBSI Standard and Radiomics Function Feature Correspondences.
Version History
Introduced in R2023bCompute texture features for multiple regions of interest (ROIs) in a medical image or medical volume.
Compute one or more types of texture features by specifying one or more options in the
name-value argument Type.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)