Analyze Vertebral Bone Density in Segmented CT Scan
This example shows how to perform quantitative CT analysis to measure bone volume and bone mineral density within a vertebra.
The goal of quantitative medical image analysis is to extract clinically meaningful information from an image. For example, you can use changes in tumor volume to track disease progression or evaluate a treatment. In this example, you calculate the volume and average density of a vertebra label mask in a computed tomography (CT) scan.
Download Data
This example uses a CT scan saved as a folder of DICOM files. The scan is part of a data set that contains three CT volumes. Run this code to download the data set from the MathWorks® website as a ZIP file and unzip the file. The total size of the data set is approximately 81 MB.
zipFile = matlab.internal.examples.downloadSupportFile("medical","MedicalVolumeDICOMData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
After you run the code to download the data, the dataFolder folder contains the downloaded and unzipped data.
dataFolder = fullfile(filepath,"MedicalVolumeDICOMData","LungCT01");
Load Data
Load the CT volume as a medicalVolume object. The Voxels property contains the image intensity values. If the file metadata specifies the rescale intercept and slope, medicalVolume automatically rescales the voxel intensities to the specified units. In this example, the rescaled CT intensity values are in Hounsfield units (HU), which is a standard intensity unit for CT. The VoxelSpacing and SpatialUnits property values specify that the size of each voxel is 0.73-by-0.73-by-2.5 mm.
medVolCT = medicalVolume(dataFolder)
medVolCT =
medicalVolume with properties:
Voxels: [512×512×88 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [0.7285 0.7285 2.5000]
NormalVector: [0 0 1]
NumCoronalSlices: 512
NumSagittalSlices: 512
NumTransverseSlices: 88
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "CT"
WindowCenters: [88×1 double]
WindowWidths: [88×1 double]
This example also uses a segmentation label mask of one vertebra, which is attached to this example as a supporting file. The mask is limited to the vertebral body, excluding the spinous processes. Load the mask as a medicalVolume object. For an example of how to segment medical image volumes in the Medical Image Labeler app, see Label 3-D Medical Image Using Medical Image Labeler.
medVolLabel = medicalVolume("LungCT01_vertebra.nii.gz");Display the CT volume with the vertebra segmentation mask as an overlay. The volshow function uses the spatial properties of medVolCT to display the volume in the patient coordinate system. The axes display indicators label the inferior/superior (S), left/right (L), and anterior/posterior (P) anatomical axes.
volshow(medVolCT, ... OverlayData=medVolLabel.Voxels, ... RenderingStyle="SlicePlanes", ... OverlayAlpha=0.5)
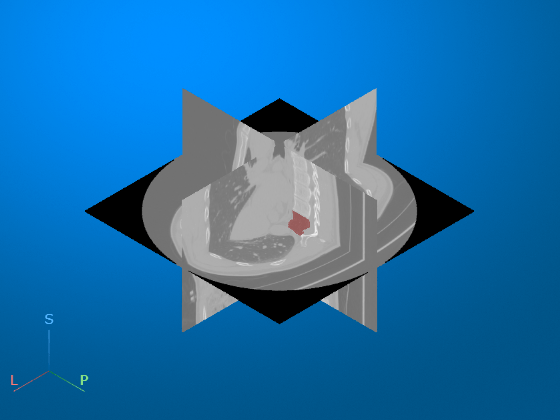
Get Image Region Properties
Calculate the volume, in voxels, and mean intensity, in Hounsfield units (HU), within the vertebra mask.
stats = regionprops3(medVolLabel.Voxels,medVolCT.Voxels,"Volume","MeanIntensity");
Calculate Bone Volume in World Units
Extract the number of voxels in the mask from the region properties structure.
numVoxels = stats.Volume;
To calculate the volume of each voxel, in cubic centimeters, convert the voxel spacing from mm to cm, and then multiply the spacing in all three directions.
dv = prod(0.1*medVolCT.VoxelSpacing);
Calculate the total volume of the vertebra, in cubic centimeters.
boneVolume = numVoxels.*dv
boneVolume = 25.4156
Calculate Bone Mineral Density
Extract the mean voxel intensity from the region properties structure. For this example, the intensity values are in Hounsfield units.
meanHU = stats.MeanIntensity;
Convert the mean intensity, in HU, to hydroxyapatite equivalent density, in grams per cubic centimeter, using this linear calibration equation [1].
boneDensity = 0.0052 + 0.0008*meanHU
boneDensity = 0.2242
References
[1] Turbucz, Mate, Agoston Jakab Pokorni, György Szőke, Zoltan Hoffer, Rita Maria Kiss, Aron Lazary, and Peter Endre Eltes. “Development and Validation of Two Intact Lumbar Spine Finite Element Models for In Silico Investigations: Comparison of the Bone Modelling Approaches.” Applied Sciences 12, no. 20 (January 2022): 10256. https://doi.org/10.3390/app122010256.
See Also
medicalVolume | regionprops3 | volshow
