Segment and Analyze Brain MRI Scan Using AI
This example shows how to preprocess, label, postprocess, and analyze a brain MRI image using MONAI Label.
In this example, you apply the wholeBrainSeg Large UNEST segmentation model [1] from the Medical Open Network for AI (MONAI) Label library. The MONAI Label [2][3] platform provides fully automated and interactive deep learning models for segmenting radiology images, and you can connect to MONAI Label from within the Medical Image Labeler app. To achieve satisfactory results from the wholeBrainSeg Large UNEST segmentation model, you must preprocess your data by registering it to an MNI305 atlas [4], which is a standardized brain atlas commonly used in neuroimaging analysis. In this example, you register the brain MRI data to the MNI305 atlas, segment it using the MONAI Label model, and transform the labels back to MRI space to calculate regional brain volumes.
Import Brain MRI Data
Import the brain MRI scan as a medicalVolume object. The MRI volume is from the CANDI data set [5] [6], and the 2.3 MB scan is attached to this example as a supporting file.
medVolMRI = medicalVolume("brainSegData_anat.nii.gz")medVolMRI =
medicalVolume with properties:
Voxels: [256×256×128 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "coronal"
VoxelSpacing: [0.9375 0.9375 1.5000]
NormalVector: [0 -1 0]
NumCoronalSlices: 128
NumSagittalSlices: 256
NumTransverseSlices: 256
PlaneMapping: ["sagittal" "transverse" "coronal"]
Modality: "unknown"
WindowCenters: 0
WindowWidths: 0
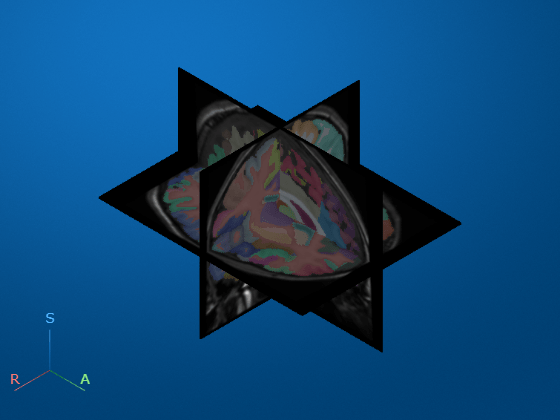
Display the brain MRI scan as 3-D slice planes.
volshow(medVolMRI,RenderingStyle="SlicePlanes")
Import Brain MNI Atlas
Import the MNI305 atlas image as a medicalVolume object. The atlas is attached to this example as a supporting file, and is approximately 2.8 MB.
atlas = medicalVolume("tpl-MNI305_T1w.nii.gz")atlas =
medicalVolume with properties:
Voxels: [172×220×156 int16]
VolumeGeometry: [1×1 medicalref3d]
SpatialUnits: "mm"
Orientation: "transverse"
VoxelSpacing: [1 1 1]
NormalVector: [0 0 1]
NumCoronalSlices: 220
NumSagittalSlices: 172
NumTransverseSlices: 156
PlaneMapping: ["sagittal" "coronal" "transverse"]
Modality: "unknown"
WindowCenters: 0
WindowWidths: 0
Display the MNI atlas image as 3-D slice planes.
volshow(atlas,RenderingStyle="SlicePlanes")
Register MRI to Atlas
Resample the brain MRI image so that it has the same voxel spacing as the brain atlas.
dataresampled = resample(medVolMRI,atlas.VolumeGeometry);
Register the resampled brain MRI scan to the atlas image using intensity-based registration. To obtain the transformation matrix that describes the registration, use the imregtform function. Specify the transformation type as "affine" so the registration includes translation, rotation, and anisotropic scaling.
[optimizer,metric] = imregconfig("multimodal"); tform = imregtform(dataresampled.Voxels,atlas.Voxels, ... "affine",optimizer,metric);
The tform object describes the transformation from the MRI image to the atlas. Apply the transformation to the resampled MRI image using imwarp. Specify the output view so the size and location of the transformed image matches the atlas.
movingRegisteredVolume = imwarp(dataresampled.Voxels,tform, ... "OutputView",imref3d(size(atlas.Voxels)));
Create a medicalVolume object that contains the transformed MRI voxel data and the spatial referencing of the MNI atlas.
medVolRegistered = medicalVolume(movingRegisteredVolume,atlas.VolumeGeometry);
Load Registered MRI into Medical Image Labeler
In this section you load the MRI scan, which has been transformed into MNI space, into the Medical Image Labeler app and perform segmentation using a MONAI Label deep learning network.
Open Medical Image Labeler
Open the Medical Image Labeler app from the Apps tab on the MATLAB® Toolstrip, under Image Processing and Computer Vision. You can also load the app by using the medicalImageLabeler command.
Create New Volume Labeling Session
To start a new 3-D labeling session, on the app toolstrip, click New Session and select New Volume Session (3-D). In the Create a new session folder dialog box, specify a location in which to save the new session folder by entering a path, or select Browse and navigate to your desired location. In the New Session Folder dialog box, specify a name for the folder for this labeling session. Then, select Create Session.
Import MRI Volume
To load the registered MRI volume into the app, on the app toolstrip, click Import, and, under Data, select medicalVolume Object. In the dialog box that opens, select medVolRegistered and click OK.
The registered MRI volume appears in the app.
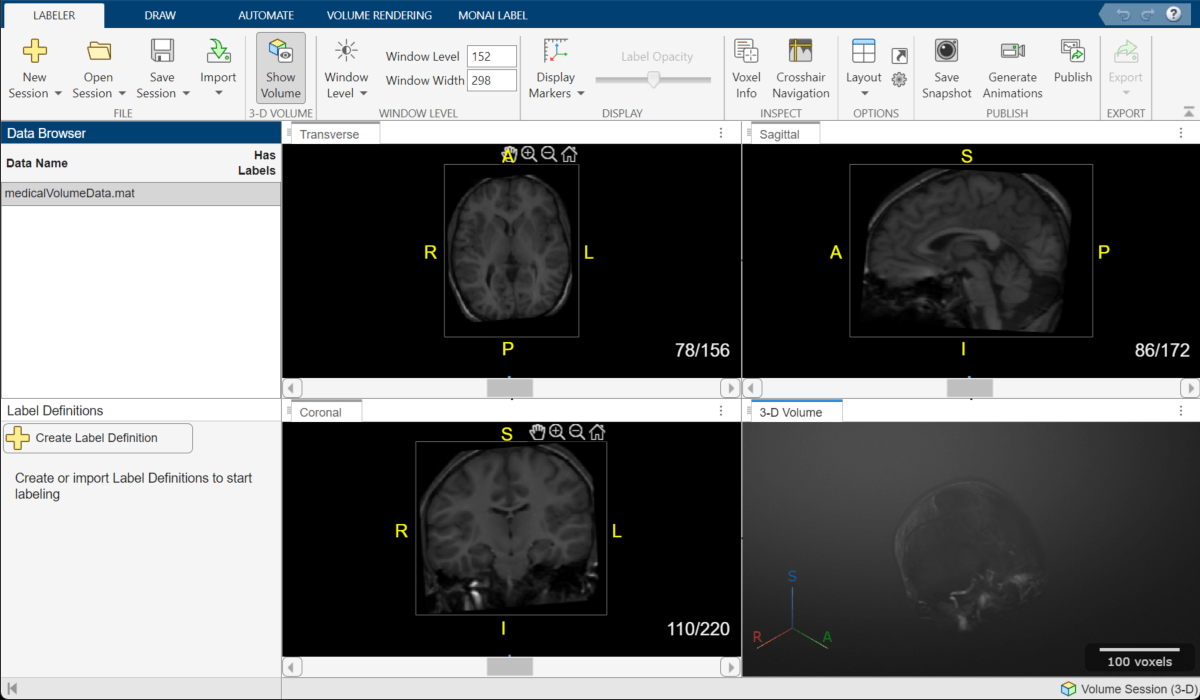
Segment MRI Volume
Start MONAI Label
On the MONAI Label tab of the app toolstrip, select Start MONAI Label > On the local machine. If this is the first time you are connecting to a local server, Medical Image Labeler prompts you to install the Medical Imaging Toolbox Interface for MONAI Label Library support package, which includes the MONAI Label software. Follow the link in the dialog box to open the Add-On Explorer. To learn more about connecting to MONAI Label, see Get Started with MONAI Label in Medical Image Labeler. For more information about installing add-ons, see Get and Manage Add-Ons. When you install the support package, MATLAB downloads the MONAI Label models, which requires an internet connection.
Once the installation is complete, try starting MONAI Label again. You do not need to restart the app.
Choose Brain MRI Segmentation Model
To select the brain segmentation model, on the MONAI Label tab of the app toolstrip, from the Deep Learning Models gallery, select the wholeBrainSeg Large UNEST segmentation model.
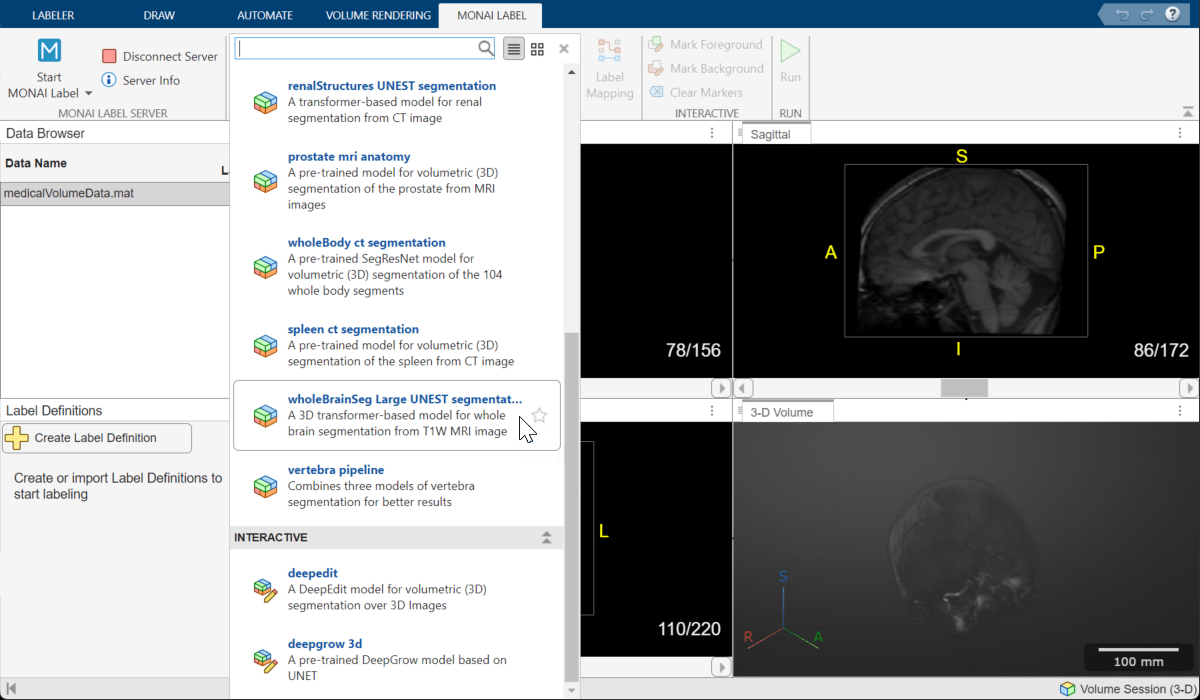
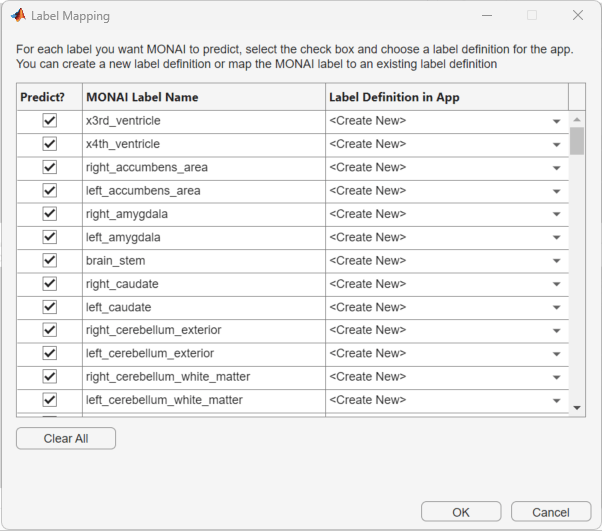
Perform Label Mapping
Use the Label Mapping dialog box to map labels predicted by the MONAI Label model to labels in the Label Definitions pane of the Medical Image Labeler app. In the dialog box, click Select All to pick all the labels. In the Label Definition in App column, the default <Create New> option enables the app to create new label definitions. Click OK to save your choices. The new labels appear in the Label Definitions pane of the app window.
Run the Model
In the app toolstrip, select Run. When the model finishes, the slice planes update to show the predicted labels. Overall, the results look satisfactory. This example uses the initial labels, and removes noise near the image boundary during postprocessing. For your data and application, you can choose to manually clean labels inside the brain using any of the manual or semi-automated drawing tools in the app.
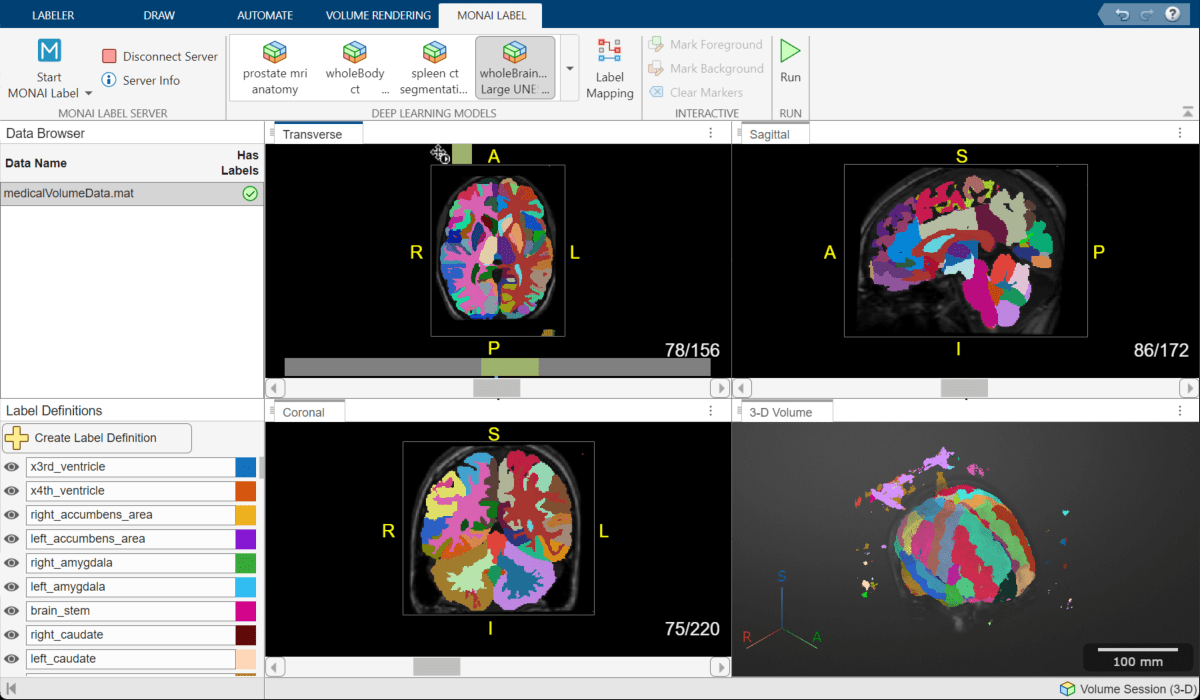
Export Ground Truth
From the Labeler tab of the app toolstrip, select Export, and, under Ground Truth, select To File. Specify the location to save the ground truth using the Export Ground Truth dialog box, and click Save. The app exports a groundTruthMedical object in the MAT file format.
Postprocess Results
Import Ground Truth
This example creates a new groundTruthMedical object that simulates you exporting results from the app to your local machine. Create the ground truth object using the createGroundTruthMed helper function, which is defined at the end of this example.
gTruthMed = createGroundTruthMed;
Alternatively, if you complete the previous sections and export your own groundTruthMedical object to the file location gTruthFileName, load the object using this code.
load(gTruthFileName)
Import the label data from the ground truth as a medicalVolume object.
labelVol = medicalVolume(gTruthMed.LabelData);
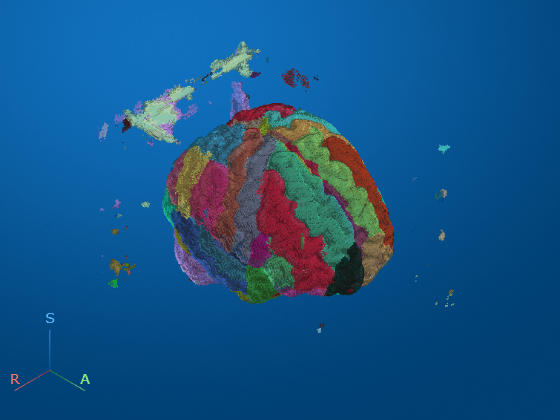
Display the registered brain MRI image as fully transparent, with the MONAI-predicted labels as an opaque overlay. As expected, the MRI data and labels are aligned.
volshow(medVolRegistered, ... alphamap=0, ... OverlayData=labelVol.Voxels, ... OverlayAlpha=0.5);
Remove the noisy voxels at the edge of the slices by using the filterNoisyVoxels helper function defined at the end of the example. The helper function loops through each label definition, and keeps only the largest connected component for that label.
numLabels = size(gTruthMed.LabelDefinitions,1); labelVolFilt = filterNoisyVoxels(labelVol,numLabels);
Display the postprocessed labels. As desired, the noisy voxels have been removed.
volshow(medVolRegistered, ... alphamap=0, ... OverlayData=labelVolFilt.Voxels, ... OverlayAlpha=0.5)

You can more closely inspect the internal postprocessed labels using the slice planes rendering style.
volshow(medVolRegistered, ... RenderingStyle="SlicePlanes", ... OverlayData=labelVolFilt.Voxels)
Transform Labels Back to MRI Space from MNI Space
The labelVolFilt volume is in the MNI space. To transform the labels back to the original brain MRI space, apply the inverse of tform to the labels. Specify the interpolation method as "nearest", which is the appropriate value for categorical label data to prevent rounding artifacts at the edges of label regions.
tformInv = invert(tform); labelVolMRI = imwarp(labelVolFilt.Voxels,tformInv,"nearest", ... OutputView=imref3d(size(dataresampled.Voxels)));
Create a medicalVolume object containing the labels in MRI space with the same spatial referencing as the unregistered, resampled Brain MRI image.
medVolLabelsMRI = medicalVolume(labelVolMRI,dataresampled.VolumeGeometry);
Resample the labels back to the original voxel spacing of the MRI scan.
medVolLabelsMRI = resample(medVolLabelsMRI,medVolMRI.VolumeGeometry,Method="nearest");Display the original MRI scan with the transformed, resampled labels as an overlay.
volshow(medVolMRI, ... RenderingStyle="SlicePlanes", ... OverlayData=medVolLabelsMRI.Voxels)
Analyze Segmented MRI Volume
Calculate the volume of different brain structures and the overall brain volume. First, you must get the label names, which are stored in the LabelDefinitions field of the groundTruthMedical object.
labelDefs = gTruthMed.LabelDefinitions
labelDefs=132×3 table
Name LabelColor PixelLabelID
_______________________________ __________________________ ____________
"x3rd_ventricle" 0.0667 0.4431 0.7451 1
"x4th_ventricle" 0.8667 0.3294 0 2
"right_accumbens_area" 0.9294 0.6941 0.1255 3
"left_accumbens_area" 0.5216 0.0863 0.8196 4
"right_amygdala" 0.2314 0.6667 0.1961 5
"left_amygdala" 0.1843 0.7451 0.9373 6
"brain_stem" 0.8196 0.0157 0.5451 7
"right_caudate" 0.3804 0.0353 0.0314 8
"left_caudate" 1 0.851 0.7176 9
"right_cerebellum_exterior" 0.702 0.5333 0.1647 10
"left_cerebellum_exterior" 0.8392 0.5804 1 11
"right_cerebellum_white_matter" 0.7961 1 0.7451 12
"left_cerebellum_white_matter" 0.0902 0.4627 0.6471 13
"right_cerebral_white_matter" 0.949 0.4039 0.7725 14
"left_cerebral_white_matter" 0.7176 0.1922 0.1725 15
"right_hippocampus" 0.7882 0.9059 1 16
⋮
Extract the label names to a new table for volume results.
results = labelDefs(:,"Name");Calculate the number of voxels in each label using the regionprops3 function.
S = regionprops3(medVolLabelsMRI.Voxels,"Volume");
numVoxels = S.Volume;To calculate the volume of each voxel, in cubic centimeters, convert the voxel spacing from mm to cm, and then multiply the spacing in all three directions. Then, calculate the volume of each label in cubic centimeters.
dv = prod(0.1*medVolMRI.VoxelSpacing); volume = numVoxels.*dv;
Append the volume values to the results table.
results.("Volume (voxels)") = numVoxels; results.("Volume (ccm)")= volume
results=132×3 table
Name Volume (voxels) Volume (ccm)
_______________________________ _______________ ____________
"x3rd_ventricle" 1280 1.6875
"x4th_ventricle" 1935 2.551
"right_accumbens_area" 340 0.44824
"left_accumbens_area" 522 0.68818
"right_amygdala" 925 1.2195
"left_amygdala" 959 1.2643
"brain_stem" 14316 18.874
"right_caudate" 2825 3.7244
"left_caudate" 2616 3.4488
"right_cerebellum_exterior" 39791 52.459
"left_cerebellum_exterior" 40408 53.272
"right_cerebellum_white_matter" 13666 18.017
"left_cerebellum_white_matter" 15885 20.942
"right_cerebral_white_matter" 1.6616e+05 219.06
"left_cerebral_white_matter" 1.688e+05 222.54
"right_hippocampus" 3202 4.2214
⋮
Calculate the total brain volume, in cubic centimeters. The total brain volume is 1446 cubic cm, which is in the normal range for human brain volume [7].
totalBrainVol = sum(volume)
totalBrainVol = 1.4462e+03
Supporting Function
The filterNoisyVoxels helper function removes small clusters of noisy label voxels using these steps:
While looping through each of the labels, create a binary mask for the current label.
Find the connected components in the mask by using the
bwconncompfunction with a connectivity value of6.Calculate the number of voxels in each connected component using the
numelfunction.Determine the indices of voxels in the connected component with the most voxels.
Set all label values for voxels outside the largest connected component to
0, which is the background. This step removes the small noisy voxels.
function labelVolFilt = filterNoisyVoxels(labelVol,numLabels) labelVolFilt = labelVol; for i = 1:numLabels maskLabel_i = labelVol.Voxels==i; CC = bwconncomp(maskLabel_i,6); numPixels = cellfun(@numel,CC.PixelIdxList); [~,idx] = max(numPixels); maskLabel_i(CC.PixelIdxList{idx}) = 0; labelVolFilt.Voxels(maskLabel_i==1) = 0; end end
The createGroundTruthMed helper function creates a groundTruthMedical object that points to the label results on your local machine. The files used to create the ground truth object are all attached to this example as supporting files. You can also create them yourself by following the processes in the Import Brain MRI Data, Import Brain MNI Atlas, Register MRI to Atlas, Load Registered MRI into Medical Image Labeler, and Segment MRI Volume sections of this example, and exporting a groundTruthMedical object from the app.
function gTruthMed = createGroundTruthMed dataFile = {"registeredVol.nii.gz"}; dataSource = medical.labeler.loading.VolumeSource(dataFile); labelData = "medicalVolumeData.nii"; load("labelDefinitions.mat"); gTruthMed = groundTruthMedical(dataSource,labelDefinitions,labelData); end
References
[1] Yu, Xin, Qi Yang, Yinchi Zhou, Leon Y. Cai, Riqiang Gao, Ho Hin Lee, Thomas Li, et al. “UNesT: Local Spatial Representation Learning with Hierarchical Transformer for Efficient Medical Segmentation.” Medical Image Analysis 90 (December 1, 2023): 102939. https://doi.org/10.1016/j.media.2023.102939.
[2] Diaz-Pinto, Andres, Sachidanand Alle, Vishwesh Nath, Yucheng Tang, Alvin Ihsani, Muhammad Asad, Fernando Pérez-García, et al. “MONAI Label: A Framework for AI-Assisted Interactive Labeling of 3D Medical Images,” 2022. https://doi.org/10.48550/ARXIV.2203.12362.
[3] Diaz-Pinto, Andres, Pritesh Mehta, Sachidanand Alle, Muhammad Asad, Richard Brown, Vishwesh Nath, Alvin Ihsani, et al. “DeepEdit: Deep Editable Learning for Interactive Segmentation of 3D Medical Images.” In Data Augmentation, Labelling, and Imperfections, edited by Hien V. Nguyen, Sharon X. Huang, and Yuan Xue, 11–21. Lecture Notes in Computer Science. Cham: Springer Nature Switzerland, 2022. https://doi.org/10.1007/978-3-031-17027-0_2.
[4] Evans, A.C., D.L. Collins, S.R. Mills, E.D. Brown, R.L. Kelly, and T.M. Peters. “3D Statistical Neuroanatomical Models from 305 MRI Volumes.” In 1993 IEEE Conference Record Nuclear Science Symposium and Medical Imaging Conference, 1813–17. San Francisco, CA, USA: IEEE, 1993. https://doi.org/10.1109/NSSMIC.1993.373602.
[5] “NITRC: CANDI Share: Schizophrenia Bulletin 2008: Tool/Resource Info.” Accessed October 17, 2022. https://www.nitrc.org/projects/cs_schizbull08/.
[6] Frazier, J. A., S. M. Hodge, J. L. Breeze, A. J. Giuliano, J. E. Terry, C. M. Moore, D. N. Kennedy, et al. “Diagnostic and Sex Effects on Limbic Volumes in Early-Onset Bipolar Disorder and Schizophrenia.” Schizophrenia Bulletin 34, no. 1 (October 27, 2007): 37–46. https://doi.org/10.1093/schbul/sbm120.
[7] Kijonka, Marek, Damian Borys, Krzysztof Psiuk-Maksymowicz, Kamil Gorczewski, Piotr Wojcieszek, Bartosz Kossowski, Artur Marchewka, Andrzej Swierniak, Maria Sokol, and Barbara Bobek-Billewicz. “Whole Brain and Cranial Size Adjustments in Volumetric Brain Analyses of Sex- and Age-Related Trends.” Frontiers in Neuroscience 14 (April 3, 2020). https://doi.org/10.3389/fnins.2020.00278.