Brain MRI Segmentation Using Pretrained 3-D U-Net Network
This example shows how to segment a brain MRI using a deep neural network.
Segmentation of brain scans enables the visualization of individual brain structures. Brain segmentation is also commonly used for quantitative volumetric and shape analyses to characterize healthy and diseased populations. Manual segmentation by clinical experts is considered the highest standard in segmentation. However, the process is extremely time-consuming and not practical for labeling large data sets. Additionally, labeling requires expertise in neuroanatomy and is prone to errors and limitations in interrater and intrarater reproducibility. Trained segmentation algorithms, such as convolutional neural networks, have the potential to automate the labeling of large clinical data sets.
In this example, you use the pretrained SynthSeg neural network [1], a 3-D U-Net for brain MRI segmentation. SynthSeg can be used to segment brain scans of any contrast and resolution without retraining or fine-tuning. SynthSeg is also robust to a wide array of subject populations, from young and healthy to aging and diseased subjects, and a wide array of scan conditions, such as white matter lesions, with or without preprocessing, including bias field corruption, skull stripping, intensity normalization, and template registration.
Download Brain MRI and Label Data
This example uses a subset of the CANDI data set [2] [3]. The subset consists of a brain MRI volume and the corresponding ground truth label volume for one patient. Both files are in the NIfTI file format. The total size of the data files is ~2.5 MB.
Run this code to download the dataset from the MathWorks® website and unzip the downloaded folder.
zipFile = matlab.internal.examples.downloadSupportFile("image","data/brainSegData.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath)
The dataDir folder contains the downloaded and unzipped dataset.
dataDir = fullfile(filepath,"brainSegData");Download and Load Pretrained Network
Download the pretrained network using downloadTrainedNetwork helper function. The helper function is attached to this example as a supporting file.
trainedBrainCANDINetwork_url = "https://www.mathworks.com/supportfiles/"+ ... "image/data/trainedSynthSegModel.zip"; downloadTrainedNetwork(trainedBrainCANDINetwork_url,dataDir)
Load the pretrained network using the importNetworkFromTensorFlow (Deep Learning Toolbox) function. The importNetworkFromTensorFlow function requires the Deep Learning Toolbox™ Converter for TensorFlow Models support package. If this support package is not installed, then the function provides a download link.
net = importNetworkFromTensorFlow(fullfile(dataDir,"trainedSynthSegModel"))Importing the saved model... Translating the model, this may take a few minutes... Finished translation. Assembling network... Import finished.
net =
dlnetwork with properties:
Layers: [60×1 nnet.cnn.layer.Layer]
Connections: [63×2 table]
Learnables: [56×3 table]
State: [18×3 table]
InputNames: {'unet_input'}
OutputNames: {'unet_prediction'}
Initialized: 1
View summary with summary.
Load Test Data
Read the metadata from the brain MRI volume by using the niftiinfo function. Read the brain MRI volume by using the niftiread function.
imFile = fullfile(dataDir,"anat.nii.gz");
metaData = niftiinfo(imFile);
vol = niftiread(metaData);In this example, you segment the brain into 32 classes corresponding to anatomical structures. Read the names and numeric identifiers for each class label by using the getBrainCANDISegmentationLabels helper function. The helper function is attached to this example as a supporting file.
labelDirs = fullfile(dataDir,"groundTruth");
[classNames,labelIDs] = getBrainCANDISegmentationLabels;Preprocess Test Data
Preprocess the MRI volume by using the preProcessBrainCANDIData helper function. The helper function is attached to this example as a supporting file. The helper function performs these steps:
Resampling — If
resampleistrue, resample the data to the isotropic voxel size 1-by-1-by-1 mm. By default,resampleis false and the function does not perform resampling. To test the pretrained network on images with a different voxel size, setresampletotrueif the input is not isotropic.Alignment — Rotate the volume to a standardized RAS orientation.
Cropping — Crop the volume to a maximum size of 192 voxels in each dimension.
Normalization — Normalize the intensity values of the volume to values in the range [0, 1], which improves the contrast.
resample = false; cropSize = 192; [volProc,cropIdx,imSize] = preProcessBrainCANDIData(vol,metaData,cropSize,resample); inputSize = size(volProc);
Convert the preprocessed MRI volume into a formatted deep learning array with the SSSCB (spatial, spatial, spatial, channel, batch) format by using dlarray (Deep Learning Toolbox).
volDL = dlarray(volProc,"SSSCB");Predict Using Test Data
Predict Network Output
Predict the segmentation output for the preprocessed MRI volume. The segmentation output predictIm contains 32 channels corresponding to the segmentation label classes, such as "background", "leftCerebralCortex", "rightThalamus". The predictIm output assigns confidence scores to each voxel for every class. The confidence scores reflect the likelihood of the voxel being part of the corresponding class. This prediction is different from the final semantic segmentation output, which assigns each voxel to exactly one class.
predictIm = predict(net,volDL);
Test Time Augmentation
This example uses test time augmentation to improve segmentation accuracy. In general, augmentation applies random transformations to an image to increase the variability of a data set. You can use augmentation before network training to increase the size of the training data set. Test time augmentation applies random transformations to test images to create multiple versions of the test image. You can then pass each version of the test image to the network for prediction. The network calculates the overall segmentation result as the average prediction for all versions of the test image. Test time augmentation improves segmentation accuracy by averaging out random errors in the individual network predictions.
By default, this example flips the MRI volume in the left-right direction, resulting in a flipped volume flippedData. The network output for the flipped volume is flipPredictIm. Set flipVal to false to skip the test time augmentation and speed up prediction.
flipVal =true; if flipVal flippedData = fliplr(volProc); flippedData = flip(flippedData,2); flippedData = flip(flippedData,1); flippedData = dlarray(flippedData,"SSSCB"); flipPredictIm = predict(net,flippedData); else flipPredictIm = []; end
Postprocess Segmentation Prediction
To get the final segmentation maps, postprocess the network output by using the postProcessBrainCANDIData helper function. The helper function is attached to this example as a supporting file. The postProcessBrainCANDIData function performs these steps:
Smoothing — Apply a 3-D Gaussian smoothing filter to reduce noise in the predicted segmentation masks.
Morphological Filtering — Keep only the largest connected component of predicted segmentation masks to remove additional noise.
Segmentation — Assign each voxel to the label class with the greatest confidence score for that voxel.
Resizing — Resize the segmentation map to the original input volume size. Resizing the label image allows you to visualize the labels as an overlay on the grayscale MRI volume.
Alignment — Rotate the segmentation map back to the orientation of the original input MRI volume.
The final segmentation result, predictedSegMaps, is a 3-D categorical array the same size as the original input volume. Each element corresponds to one voxel and has one categorical label.
predictedSegMaps = postProcessBrainCANDIData(predictIm,flipPredictIm,imSize, ...
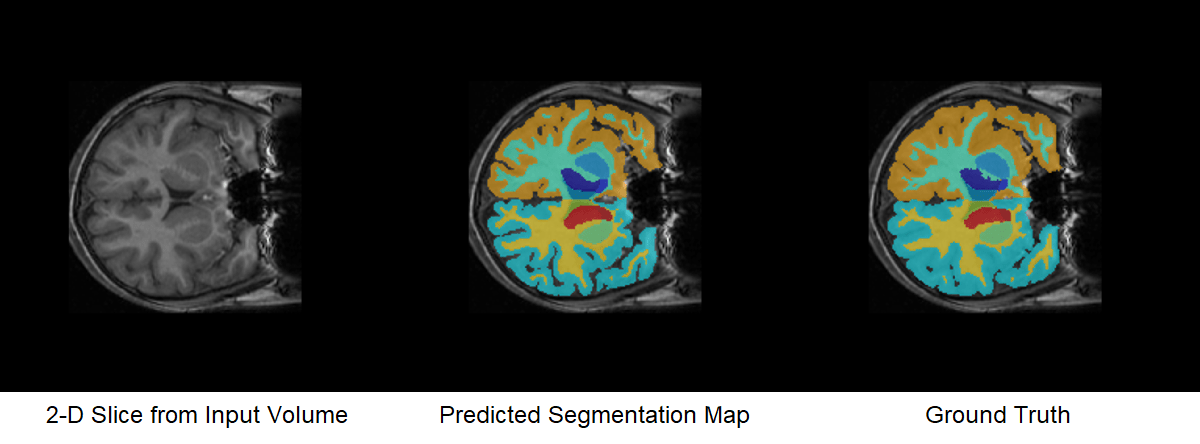
cropIdx,metaData,classNames,labelIDs);Overlay a slice from the predicted segmentation map on a corresponding slice from the input volume using the labeloverlay function. Include all the brain structure labels except the background label.
sliceIdx = 80;
testSlice = rescale(vol(:,:,sliceIdx));
predSegMap = predictedSegMaps(:,:,sliceIdx);
B = labeloverlay(testSlice,predSegMap,"IncludedLabels",2:32);
figure
montage({testSlice,B})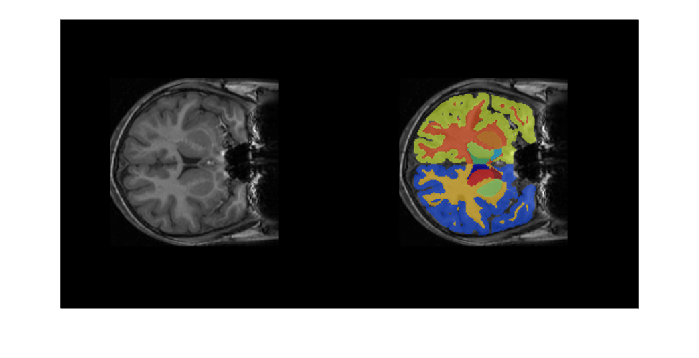
Quantify Segmentation Accuracy
Measure the segmentation accuracy by comparing the predicted segmentation labels with the ground truth labels drawn by clinical experts.
Create a pixelLabelDatastore (Computer Vision Toolbox) to store the labels. Because the NIfTI file format is a nonstandard image format, you must use a NIfTI file reader to read the pixel label data. You can use the helper NIfTI file reader, niftiReader, defined at the bottom of this example.
pxds = pixelLabelDatastore(labelDirs,classNames,labelIDs,FileExtensions=".gz",... ReadFcn=@(X)uint8(niftiread(X)));
Read the ground truth labels from the pixel label datastore.
groundTruthLabel = read(pxds);
groundTruthLabel = groundTruthLabel{1};Measure the segmentation accuracy using the dice function. This function computes the Dice index between the predicted and ground truth segmentations.
diceResult = zeros(length(classNames),1); for j = 1:length(classNames) diceResult(j)= dice(groundTruthLabel==classNames(j),... predictedSegMaps==classNames(j)); end
Calculate the average Dice index across all labels for the MRI volume.
meanDiceScore = mean(diceResult);
disp("Average Dice score across all labels = " +num2str(meanDiceScore))Average Dice score across all labels = 0.7579
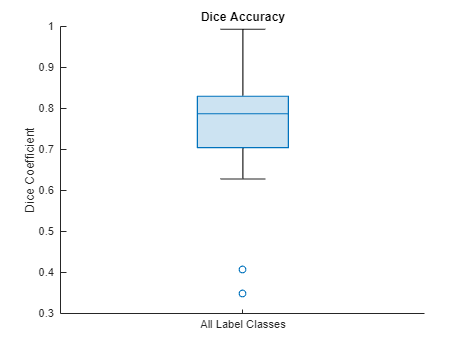
Visualize statistics about the Dice indices across all the label classes as a box chart. The middle blue line in the plot shows the median Dice index. The upper and lower bounds of the blue box indicate the 25th and 75th percentiles, respectively. Black whiskers extend to the most extreme data points that are not outliers.
figure boxchart(diceResult) title("Dice Accuracy") xticklabels("All Label Classes") ylabel("Dice Coefficient")
References
[1] Billot, Benjamin, Douglas N. Greve, Oula Puonti, Axel Thielscher, Koen Van Leemput, Bruce Fischl, Adrian V. Dalca, and Juan Eugenio Iglesias. “SynthSeg: Domain Randomisation for Segmentation of Brain Scans of Any Contrast and Resolution.” ArXiv:2107.09559 [Cs, Eess], December 21, 2021. https://arxiv.org/abs/2107.09559.
[2] “NITRC: CANDI Share: Schizophrenia Bulletin 2008: Tool/Resource Info.” Accessed October 17, 2022. https://www.nitrc.org/projects/cs_schizbull08/.
[3] Frazier, J. A., S. M. Hodge, J. L. Breeze, A. J. Giuliano, J. E. Terry, C. M. Moore, D. N. Kennedy, et al. “Diagnostic and Sex Effects on Limbic Volumes in Early-Onset Bipolar Disorder and Schizophrenia.” Schizophrenia Bulletin 34, no. 1 (October 27, 2007): 37–46. https://doi.org/10.1093/schbul/sbm120.
See Also
niftiread | importNetworkFromTensorFlow (Deep Learning Toolbox) | predict (Deep Learning Toolbox) | pixelLabelDatastore (Computer Vision Toolbox) | dice | boxchart