Get Started with Medical Segment Anything Model for Medical Image Segmentation
Medical Segment Anything Model (MedSAM) is a deep learning algorithm for the easy, interactive segmentation of 2-D medical images. You can use MedSAM to segment objects in several medical imaging modalities, without needing to retrain the model for your data set. MedSAM builds upon the foundation of the Segment Anything Model (SAM).
SAM is a pretrained image segmentation model that you can use along with visual prompts to accurately segment objects within images without further training. The visual prompts are inputs to the model that characterize the object you want to segment. For example, you can provide bounding boxes around the object you want to segment. SAM is also a zero-shot segmentation model. You do not need to provide examples of prompts and their corresponding segmentation masks to SAM.
MedSAM extends the applicability of SAM to the medical domain, enabling accurate and efficient segmentation of medical images. While SAM is trained on the SA-1B Dataset, MedSAM is trained on a large-scale medical image data set with over 1.5 million image-mask pairs, covering several imaging modalities and cancer types.
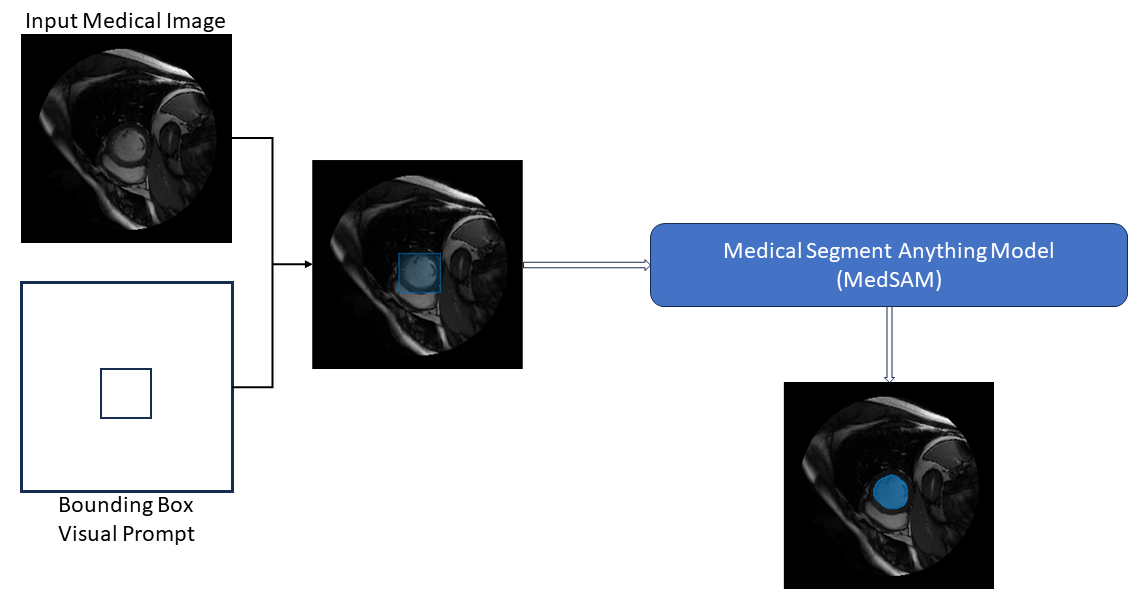
The architecture consists of these key components:
Image Encoder — The image encoder generates image embeddings. An image embedding is a feature representation of an image that encodes the semantics of its contents. The image encoder analyze the image and generate multiple features. All these features together make up the image embedding.
Prompt Encoder — The prompt encoder transforms user-provided prompts (such as bounding boxes) into feature representations. Essentially, it encodes the spatial information from the prompts, enabling MedSAM to understand the context of the object you want to segment, and guide its segmentation process.
Mask Decoder — The mask decoder generates segmentation masks using the image embeddings and prompt embeddings.
You can perform medical image segmentation using the Medical Imaging Toolbox™ Model for Medical Segment Anything Model.
Install Support Package
You can install the Medical Imaging Toolbox Model for Medical Segment Anything Model from the Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons. The support package also requires Deep Learning Toolbox™ and Computer Vision Toolbox™. Processing image data on a GPU requires a supported GPU device and Parallel Computing Toolbox™.
Segment Medical Images Using MedSAM
Use this process to segment medical images using MedSAM.
Load a medical image into the workspace and store the image size in the
imageSizevariable.zipFile = matlab.internal.examples.downloadSupportFile("medical","CardiacMRI.zip"); filepath = fileparts(zipFile); unzip(zipFile,filepath) imageDir = fullfile(filepath,"Cardiac MRI"); medImg = medicalImage(fullfile(imageDir,"images","SC-HF-I-01","SC-HF-I-01_rawdcm_099.dcm")); I = medImg.Pixels; I = mat2gray(I);
MedSAM supports 2-D grayscale images, 2-D RGB images, a batch of 2-D grayscale images, or a batch of 2-D RGB images. If you are segmenting a batch of B images, load them all into the workspace. All the images in the batch must have the same size.
Create a
medicalSegmentAnythingModelobject to configure the pretrained MedSAM.medsam = medicalSegmentAnythingModel;
Extract the image embeddings of your image or batch of B images using the
extractEmbeddingsobject function.embeddings = extractEmbeddings(medsam,I);
If you are working with a single image,
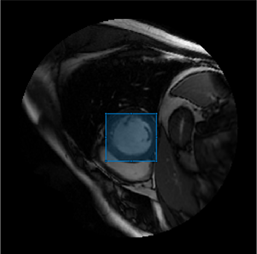
embeddingshas a size of 64-by-64-256. If you are working with a batch of B images,embeddingshas a size of 64-by-64-256-by-B.Specify the visual prompt for segmentation as a bounding box for the object you want to segment. Even if you have extracted embeddings for a batch of images, you can segment only one image at a time. If you are segmenting a batch of images, select which image to segment and specify the visual prompt.
For a single image,
I, you can specify the visual prompt programmatically or interactively.Programmatic Visual Prompt
boxPrompt = [100 110 50 50];
Interactive Visual Prompt
f = figure; ax = axes(f); imshow(I,[],Parent=ax); roi = drawrectangle(ax); boxPrompt = roi.Position;
For a single image,
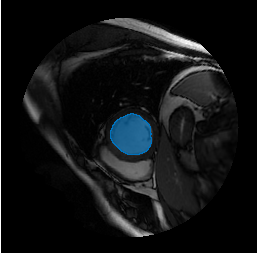
I, and its embeddings,embeddings, segment the object defined by the bounding box in the image using thesegmentObjectsFromEmbeddingsobject function.imageSize = size(I); mask = segmentObjectsFromEmbeddings(medsam,embeddings,imageSize,BoundingBox=boxPrompt);
Overlay the detected object mask on the input image, and display the image with its object mask.
imMask = insertObjectMask(I,mask); imshow(imMask)
You can interactively segment objects in medical images and in cross-sections of medical volumes using the MedSAM algorithm in the Medical Image Labeler app. To get started, see Get Started with MedSAM in Medical Image Labeler.
Applications
You can perform medical image segmentation on medical images using MedSAM at different stages in a medical imaging application. You can perform downstream tasks such segmenting detected objects for applications like organ segmentation. You can also perform radiomics analyses of the segmented objects.
References
[1] Ma, Jun, Yuting He, Feifei Li, Lin Han, Chenyu You, and Bo Wang. “Segment Anything in Medical Images.” Nature Communications 15, no. 1 (January 22, 2024): 654. https://doi.org/10.1038/s41467-024-44824-z.
See Also
medicalSegmentAnythingModel | extractEmbeddings | segmentObjectsFromEmbeddings | Medical Image
Labeler
Topics
- Interactively Segment and Analyze ROI Using MedSAM and Radiomics
- Get Started with Medical Image Labeler
- Get Started with MedSAM in Medical Image Labeler
- Get Started with MONAI Label in Medical Image Labeler