segmentObjectsFromEmbeddings
Segment objects in medical image using Medical Segment Anything Model (MedSAM) image embeddings
Since R2024b
Syntax
Description
[
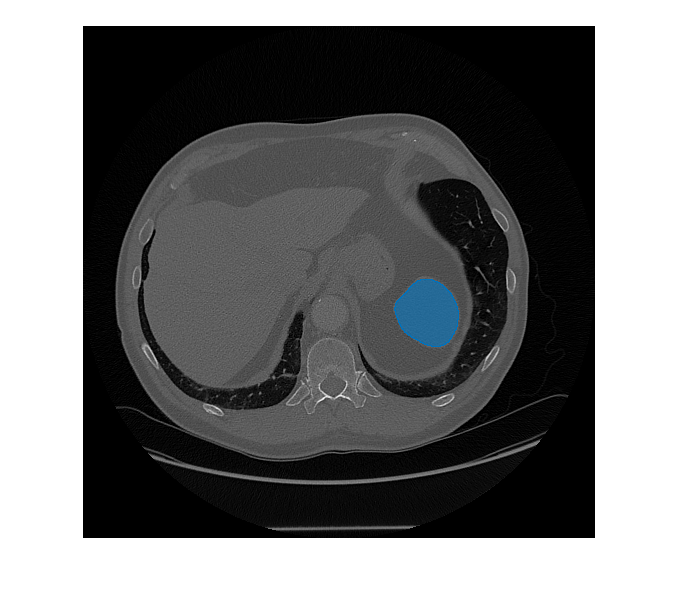
segments objects from an image of size mask,score] = segmentObjectsFromEmbeddings(medsam,embeddings,imageSize,BoundingBox=boxPrompt)imageSize using the Medical
Segment Anything Model (MedSAM) image embeddings embeddings and the
bounding box coordinates boxPrompt as a visual prompt, and returns the
predicted object mask mask and the corresponding prediction score
score.
Note
This functionality requires Deep Learning Toolbox™, Computer Vision Toolbox™, and the Medical Imaging Toolbox™ Model for Medical Segment Anything Model. You can install the Medical Imaging Toolbox Model for Medical Segment Anything Model from Add-On Explorer. For more information about installing add-ons, see Get and Manage Add-Ons.
Examples
Input Arguments
Output Arguments
Extended Capabilities
Version History
Introduced in R2024b