运行不使用并行池的 parfor 循环
此示例显示如何在没有并行池的大型集群上运行 parfor 循环。
直接在集群上运行 parfor 计算允许您使用数百个工作单元来执行 parfor 循环。当您使用此方法时,parfor 可以使用集群中所有可用的工作单元,并在循环完成后立即释放工作单元。如果您的集群不支持并行池,这种方法也很有用。然而,当您直接在集群上运行 parfor 计算时,您无法访问 DataQueue 或 Constant 对象,并且工作单元会在迭代之间重新启动,这可能会导致大量开销。
此示例重新创建了 Jammer 等人 [2] 对 ARGESIM 基准 CP2 蒙特卡罗研究 [1] 的更新。对于 CP2 蒙特卡罗研究,您可以并行仿真具有不同随机采样阻尼系数的弹簧质量阻尼器系统。
创建集群对象
创建集群对象并显示集群中可用的工作单元数量。HPCProfile 是 MATLAB® 作业调度器集群的配置文件。
cluster = parcluster("HPCProfile"); maxNumWorkers = cluster.NumWorkers; fprintf("Number of workers available: %d",maxNumWorkers)
Number of workers available: 496
定义仿真参数
设置质量弹簧系统 ODE 的仿真周期、时间间隔和初始状态。
period = [0 2]; % Use a period from 0 to 2 seconds h = 0.001; % time step t_interval = period(1):h:period(2); y0 = [0 0.1];
设置迭代次数。
nReps = 10000000;
初始化随机数生成器并创建一个从范围为 [800,1200] 的均匀分布中采样的阻尼系数数组。
rng(0); a = 800; b = 1200; d = (b-a).*rand(nReps,1)+a;
并行运行 ODE 求解并行
初始化归约操作的结果变量。
y_sum = zeros(numel(t_interval),1);
在 parfor 循环中执行 ODE 求解器来仿真具有不同阻尼系数的系统。要在集群上直接运行 parfor 计算,请将集群对象作为第二个输入参量传递给 parfor。使用归约变量来计算每个时间步骤的运动总和。
parfor(n = 1:nReps,cluster) f = @(t,y) massSpringODE(t,y,d(n)); [tOut,yOut] = ode45(f,t_interval,y0); y_sum = y_sum + yOut(:,1); end
计算系统的平均响应并绘制响应与时间的关系图。
meanY = y_sum./numel(d); plot(t_interval,meanY) title("ODE Solution of Mass-Spring System") xlabel("Time") ylabel("Motion") grid on
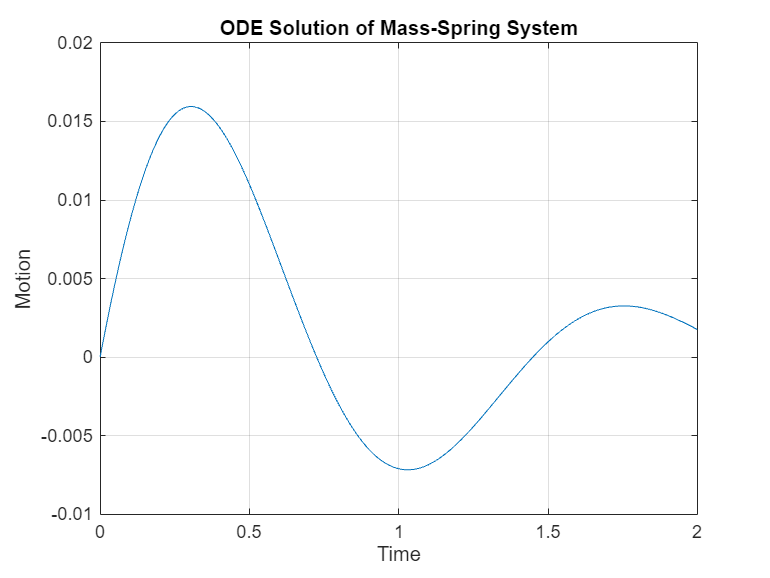
比较计算加速
比较在集群上直接运行 parfor 循环与在并行池上运行 parfor 循环的计算加速。
使用附加到此示例的 timeExecution 辅助函数来测量客户端上、在具有 496 个工作单元的并行池上以及直接在具有 496 个可用工作单元的集群上的 parfor 循环工作流的执行时间。
[serialTime,hpcPoolTime,hpcClusterTime] = timeExecution("HPCProfile",maxNumWorkers);
elapsedTimes = [serialTime hpcPoolTime hpcClusterTime];计算计算加速。
speedUp = elapsedTimes(1)./elapsedTimes;
fprintf("Speedup on cluster = %4.2f\nSpeedup on pool = %4.2f",speedUp(3),speedUp(2))Speedup on cluster = 154.11 Speedup on pool = 171.23
创建一个条形图来比较每次执行的加速情况。图显示,直接在集群上运行 parfor 循环与在并行池上运行 parfor 循环具有相似的加速。
figure; x = ["Client","Pool","Cluster"]; bar(x,speedUp); ylabel("Computational Speedup") xlabel("parfor Execution Environment") grid on
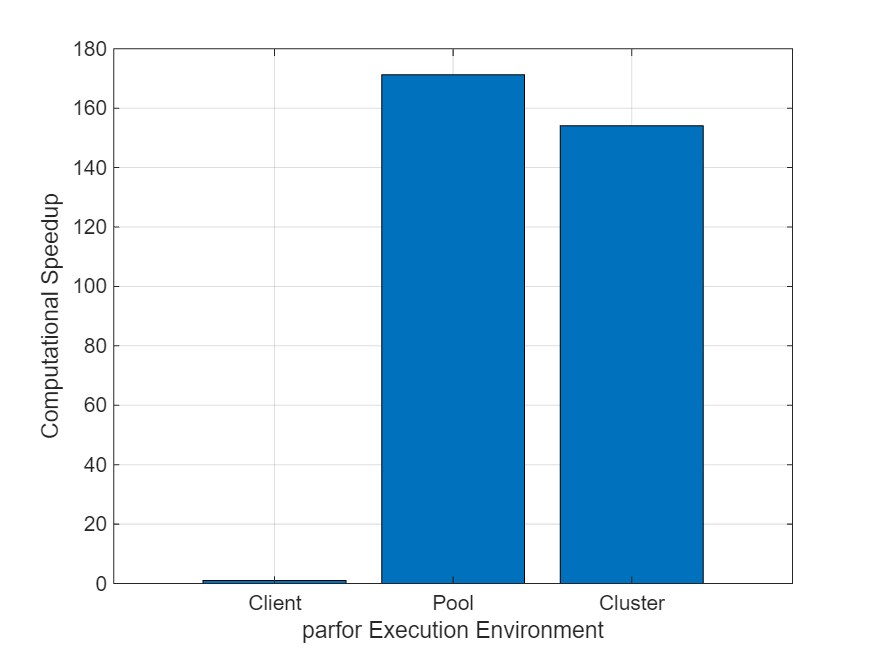
由于示例使用了 MATLAB 作业调度器集群,因此加速值相似。当您直接在 MATLAB 作业调度器集群上运行 parfor 循环时,parfor 有时可以重新使用工作单元而无需在迭代之间重新启动它们,从而减少了开销。如果直接在第三方调度器集群上运行 parfor 循环,则 parfor 会在迭代之间重新启动工作单元,这可能会导致大量开销和更低的加速值。
辅助函数
该辅助函数表示求解器使用的质量弹簧系统的 ODE。
您可以将描述弹簧质量系统 (eq1) 的微分方程重写为可以使用 ode45 求解器进行求解的一阶 ODE 系统 (eq2)。
(eq1)
(eq2)
function dy = massSpringODE(t,y0,d) k = 9000; % spring stiffness (N/m) m = 450; % mass (kg) dy = zeros(2,1); dy(1) = y0(2); dy(2) = -(d*y0(2)+k*y0(1))/m; end
参考资料
[1] Breitenecker, Felix, Gerhard Höfinger, Thorsten Pawletta, Sven Pawletta, and Rene Fink."ARGESIM Benchmark on Parallel and Distributed Simulation."Simulation News Europe SNE 17, no. 1 (2007):53-56.
[2] Jammer, David, Peter Junglas, and Sven Pawletta.“Solving ARGESIM Benchmark CP2 ’Parallel and Distributed Simulation’ with Open MPI/GSL and Matlab PCT - Monte Carlo and PDE Case Studies.”SNE Simulation Notes Europe 32, no. 4 (December 2022):211–20. https://doi.org/10.11128/sne.32.bncp2.10625.