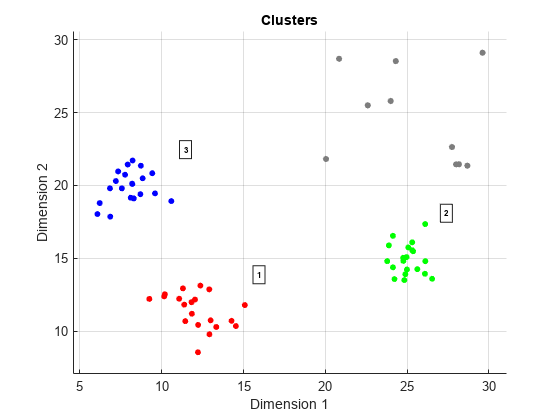
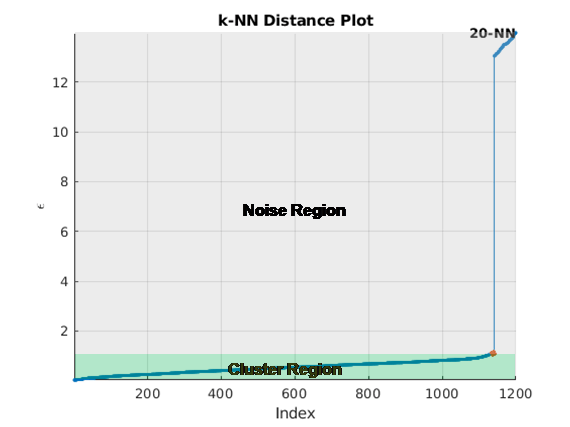
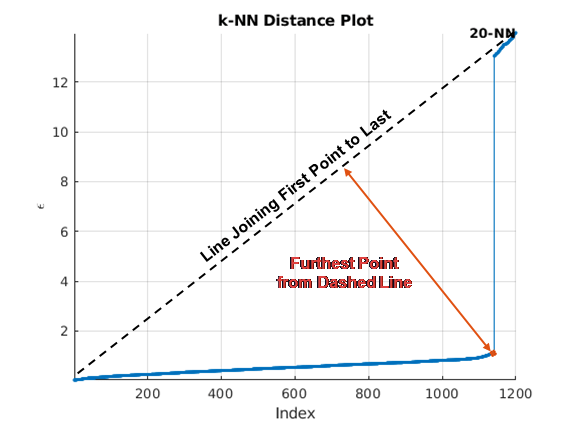
clusterDBSCAN.estimateEpsilon
Syntax
Description
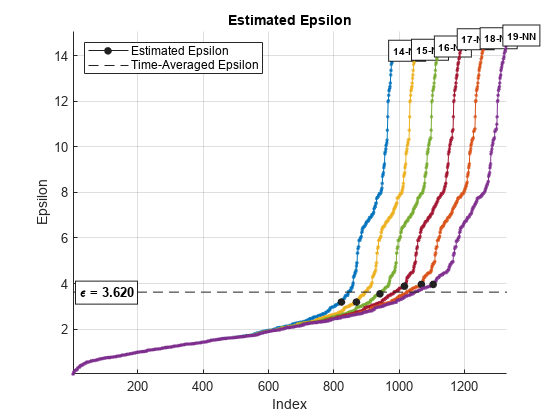
epsilon = clusterDBSCAN.estimateEpsilon(X,MinNumPoints,MaxNumPoints)epsilon,
used in the density-based spatial clustering of applications with noise (DBSCAN) algorithm.
epsilon is computed from input data X using a
k-nearest neighbor (k-NN) search.
MinNumPoints and MaxNumPoints set a range of
k-values for which epsilon is calculated. The range extends from
MinNumPoints – 1 through MaxNumPoints – 1.
k is the number of neighbors of a point, which is one less than the
number of points in a neighborhood.
clusterDBSCAN.estimateEpsilon(
displays a figure showing the k-NN search curves and the estimated
X,MinNumPoints,MaxNumPoints)epsilon. The neighborhood clustering threshold,
epsilon, is used in the density-based spatial clustering of
applications with noise (DBSCAN) algorithm. epsilon is computed from
input data X using a k-nearest neighbor
(k-NN) search. MinNumPoints and
MaxNumPoints set a range of k-values for which
epsilon is calculated. The range extends from MinNumPoints – 1 through
MaxNumPoints – 1. k is the number of neighbors
of a point, which is one less than the number of points in a neighborhood.
Examples
Input Arguments
Output Arguments
Algorithms
Extended Capabilities
Version History
Introduced in R2021a