sbionca
Compute noncompartmental analysis (NCA) parameters (requires Statistics and Machine Learning Toolbox)
Description
ncaparameters = sbionca(data,opt)data. The
options object opt defines the data columns and other
calculation options. ncaparameters is a table of calculated NCA
parameter values for each group.
Examples
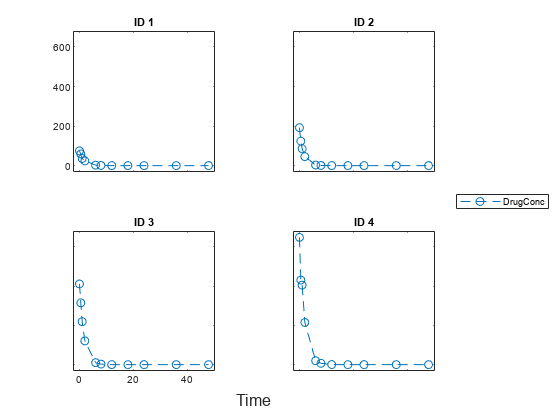
Load a synthetic data set that contains the drug concentration measurements of four individuals after an IV bolus dose.
load data1.matSet the dose amounts to NaN at time points when no dose was administered.
data1.Dose(data1.Dose(:) == 0) = NaN;
Display the data.
sbiotrellis(data1,'ID','Time','DrugConc','Marker','o','LineStyle','--');
Categorize the data columns using an NCA options object.
opt = sbioncaoptions; opt.groupColumnName = 'ID'; opt.concentrationColumnName = 'DrugConc'; opt.timeColumnName = 'Time'; opt.IVDoseColumnName = 'Dose';
Compute NCA parameters for each individual.
ncaparameters = sbionca(data1,opt);
Display the first few columns of the table. Each row of ncaparameters table represents an individual (or group), and each column lists the corresponding NCA parameter value.
ncaparameters(:,1:15)
ans=4×15 table
ID doseSchedule administrationRoute Lambda_Z R2 adjusted_R2 Num_points AUC_0_last Tlast C_max C_max_Dose T_max MRT T_half AUC_infinity
__ ____________ ___________________ ________ _______ ___________ __________ __________ _____ ______ __________ _____ ______ ______ ____________
1 {'Single'} {'IVBolus'} 0.57893 0.99991 0.9999 11 143.61 48 74.412 1.4882 0 1.5408 1.1973 143.61
2 {'Single'} {'IVBolus'} 0.66798 0.99998 0.99998 11 299.37 48 191.96 1.9196 0 1.3352 1.0377 299.37
3 {'Single'} {'IVBolus'} 0.62124 0.99999 0.99999 11 766.5 48 411.06 1.6442 0 1.4476 1.1157 766.5
4 {'Single'} {'IVBolus'} 0.58011 0.99995 0.99995 11 1301.8 48 648.33 1.2967 0 1.5721 1.1949 1301.8
You can also specify a custom time range to compute T_max and C_max within that time range, say from time = 0 to 20. You can do so by setting the C_max_ranges property as a cell array of two-element row vector.
opt.C_max_ranges = {[5.5 20]};
ncaparameters2 = sbionca(data1,opt);The function reports the T_max and C_max values within the range by adding two new columns: T_max_5_5__20 and C_max_5_5__20. Note that in the names of these two columns, the last time point is preceded by two consecutive underscores (__).
ncaparameters2.T_max_5_5__20(:)
ans = 4×1
6
6
6
6
ncaparameters2.C_max_5_5__20(:)
ans = 4×1
2.2719
3.0213
10.0233
19.9006
Similarly, you can specify a custom time range to compute the partial AUC value for each group.
opt.PartialAreas = {[0 20]};
ncaparameters3 = sbionca(data1,opt);
ncaparameters3.AUC_0__20(:)ans = 4×1
103 ×
0.1436
0.2994
0.7665
1.3017
You can also specify multiple time ranges for C_max_ranges and PartialAreas.
opt.C_max_ranges = {[0 20],[0 10],[0 15]};
opt.PartialAreas = {[0 12],[0 30]};
ncaparameters4 = sbionca(data1,opt);Input Arguments
Concentration-time data for NCA parameter computation, specified as a
table, dataset, or groupedData.
The data must contain a time column, a concentration column, and a dose column that defines dose amounts. The function ignores all data points before the first dose. Optionally, if you have data containing multiple groups of observations, you can define a group column. You can also specify a dose rate column as needed. For details, see Noncompartmental Analysis.
Example: concData
Options object to define data columns and calculation options, specified
as an NCA options object. Use sbioncaoptions to create this object and set the
options.
Example: ncaopt
Output Arguments
Calculated NCA parameters, returned as a table. For details on how the parameters are computed, see Noncompartmental Analysis.
Version History
Introduced in R2017bYou can use a groupedData object as the input data
for sbionca.
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)