Supported Files and Data Types
You can import tabular data to the SimBiology Model Analyzer app or to the MATLAB® Workspace. The supported file types are Excel® files (.xls, .xlsx), text files (.csv, .txt), and SAS® XPORT files (.xpt). You can also specify that the data is in a NONMEM® formatted file. The import process interprets the columns according to the NONMEM definitions. For more information see Support for Importing NONMEM Formatted Files.
Note
If your data set contains dosing information that is infusion data, the data set must contain the rate and not an infusion duration.
Unit Conversion
Regardless of whether unit conversion functionality is on or off, dosing in the data
file must be expressed in amounts (or as amount/time for infusion rate).
By default Unit Conversion is off, so you must ensure that units for
the data are consistent with each other. If you want to turn on unit conversion, see Unit Conversion for Imported Data.
Create Data File with SimBiology Definitions
If you are creating a file containing time course data that you want to import into SimBiology® for fitting, create the data file with the following columns:
Group column — Specify text, numeric, or categorical values. For instance, you can use this column to group multiple individuals into separate groups. You can then use this grouping or categorical information for hierarchical fits. This column is optional.
ID column — Specify text, numeric, or categorical values. The rows in the file that have the same ID column value are for the same individual. This column is optional if the measurement data comes from just one individual.
Time column — Specify monotonically increasing positive values within each ID that define the time of the dose, observation measurements, and covariate measurements.
Zero or more dosing columns — Create one dosing column for each compartment being dosed. In each column, specify positive values representing dose amounts that are added to a species. Use
NaN(not a number) to specify that no dose was applied at the specified time. In other words, specify the dose amount asNaNwhen an observation was recorded but no dose was applied.Zero or more rate columns — Specify positive values, zero, or
NaN. Zero specifies an infinite rate andNaNspecifies that no rate applies. The rate column is associated with a dosing column and defines the rate at which the dose is administered. For example, if you can specify an infusion dose in theDose1column, specify its rate in theRate1column.Zero or more observation columns — Specify numeric values or
NaN.NaNvalues define that no observation was recorded at the specified time. UseNaNfor times when a dose was applied but no observation was recorded. You can specify one observation value at a particular time for each ID. When you have replicates, specify multiple observation values for the same time point by adding more rows with the same time value. For an example, see rows 2 and 3 in the screen shot below, where CentralConc has two measurements at time = 0.Zero or more covariate columns — Specify text, numeric, or categorical values, or
NaN. Each value defines the covariate value at the specified time.NaNvalues indicate that no covariate observation was recorded at the specified time. SimBiology supports only covariates that are not time varying. For instance, see the Sex and Age columns in the example below. For an example that shows how to use categories for fitting, see Estimate Category-Specific PK Parameters for Multiple Individuals. You can also create variants from covariate columns. For details, seecreateVariants.
A screen shot of a sample data file follows.
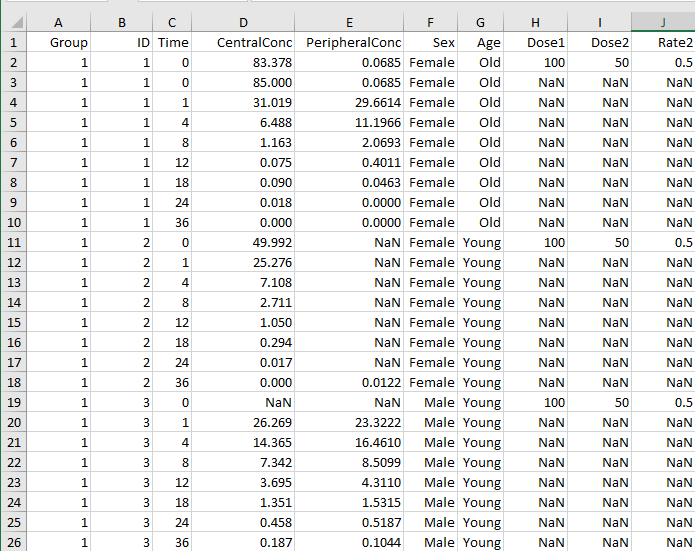
You can download the sample Excel file from the following location:
matlabroot/examples/simbio/data/sample_data_simbiology.xlsx.
matlabroot is the root directory where you installed MATLAB. You can
also enter matlabroot at the command line to see the
file path of the root directory.
Support for Importing NONMEM Formatted Files
You can specify that the data is in a NONMEM formatted file. The following table highlights the interpretation of this data in SimBiology software.
| Column Header | Interpretation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
ID | Text (character vector), numeric, or categorical values that identify the
record or group. The import process assumes that contiguous data with the same
value contains data from one individual. If the data contains non-contiguous
references to the same value, the import process assigns the second ID encountered
an indexed valued derived from the group first encountered. For example, if the ID
columns contains | ||||||||||
TIME | Monotonically increasing positive values within each group, indicating
time of observation or dose or text (character vector). The data file can specify
clock ( The following table is an example of how the import process interprets the clock values as decimal values.
If the data file also contains a | ||||||||||
DATE, DAT1, DAT2,
or DAT3 | Defines the day of the observation or the dose. The column can contain
the month as a number (
Note
| ||||||||||
DV | Numeric value of an observation. Column cannot contain Inf
or –Inf. | ||||||||||
MDV
| Defines whether a row describes an observation:
| ||||||||||
EVID | Defines the type of event described for the row in the record:
If a column contains values for dose, but
If
The import
process does not support values | ||||||||||
CMT
| Indicates which compartment is used for observation value or for dose received.
The interpretation also depends on EVID:
Note SimBiology numbers compartments starting with 1, while NONMEM numbers them
starting with 0. For instance, if a NONMEM data file contains doses and
measurements for | ||||||||||
AMT
| Positive number indicating dose. 0 or NaN
specifies no dose administered. The column cannot contain
Inf. | ||||||||||
RATE | Positive number indicating rate of infusion. 0 specifies an
infinite rate (equivalent to a bolus dose), and NaN specifies no
rate. The column cannot contain Inf. | ||||||||||
II | Positive number defining the time between doses. | ||||||||||
ADDL | When the data specifies a number of identical serial doses at specific
intervals (defined by II), ADDL specifies the
number of doses in the series excluding the initial dose. If the data specifies
II but not ADDL, then SimBiology assumes that the dosing occurs for the duration of that data record.
|
Unsupported NONMEM Definitions
The import process does not support (and therefore ignores) the rows containing the following values or definitions:
EVIDvalues 3 and 4SScolumn for specifying steady state dosesPCMTcolumn to define whether to compute a prediction for the rowCALLcolumn for calling the ERROR or the PK subroutineIf rate is specified as being less than zero, it is assumed to be zero
Supported Table Column Types in SimBiology Model Analyzer
Yon can import real and nonsparse numeric data into the SimBiology Model
Analyzer app, but not complex or sparse numeric values.
When you are
importing data from a table into the app, the app supports the following column data types:
double, char, string, cell array
of character vectors, categorical, duration,
logical, and datetime. The table that you are
importing cannot contain complex or sparse numeric values.
Support for Importing Multidimensional SimData to SimBiology Model Analyzer
When you import a multidimensional SimData array to the app, the app
flattens the SimData array and uses a single index (linear indexing) in the
corresponding datasheet. For example, if you import a 2x2x2 SimData array
A, the app creates a datasheet with 8 groups (one for each
SimData object), indexing from 1 to 8. The app still displays the
original size of the SimData array in the
Browser.