Create Pharmacokinetic Models
Ways to Create or Import Pharmacokinetic Model
To start modeling, you can:
Create a PK model using a model construction wizard that lets you specify the number of compartments, the route of administration, and the type of elimination.
Extend any model to build higher fidelity models.
Build or load your own model. Load a SimBiology® project or SBML model.
How SimBiology Models Represent Pharmacokinetic Models
The following figure compares a model as typically represented in pharmacokinetics with the same model shown in the SimBiology model diagram. For this comparison, assume that you are modeling administration of a drug using a two-compartment model with any dosing input and linear elimination kinetics. (The model structure remains the same with any dosing type.)
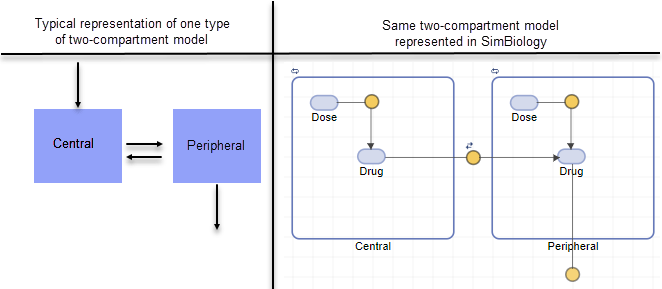
Note the following:
SimBiology represents the concentration or amount of a drug in a given compartment or volume by a species object contained within the compartment.
SimBiology represents the exchange or flow of the drug between compartments and the elimination of the drug by reactions.
SimBiology represents intercompartmental clearance by a parameter (
Q) which specifies the clearance between the compartments.SimBiology drives the dosing schedule with a combination of species (
Drugand/orDose) and reactions (Dose -> Drug), depending on whether the administration into the compartment follows bolus, zero-order, infusion, or first-order dosing kinetics. For more information on the components added and parameters estimated, see Dosing Types.
You can also view this model as a regression function, y = f(k,u),
where y is the predicted value, given values of an input
u, and parameter values k. In SimBiology the model represents f, and the model is used to generate a
regression function if y, k, and u
are identified in the model.
Dosing Types
When creating models, SimBiology creates the following model components for each compartment in the model, regardless of the dosing type:
Two species (
Drug_CompartmentNameandDose_CompartmentName) for each compartment.A reaction (
Dose_CompartmentName -> Drug_CompartmentName) for each compartment, governed by mass action kinetics.A parameter (
ka_CompartmentName) for each compartment, representing the absorption rate of the drug when absorption follows first-order kinetics. This is the forward rate parameter for theDose_CompartmentName -> Drug_CompartmentNamereaction.A parameter (
Tk0_CompartmentName) for each compartment, representing the duration of drug absorption when absorption follows zero-order kinetics.A parameter (
TLag_CompartmentName) for each compartment, representing the time lag for any dose that targets that compartment and also that is specified as having a time lag.
For dosing types that have a fixed infusion or absorption duration
(infusion and zero-order), you can use overlapping
doses. The doses are additive.
The following table describes the dosing types, the default parameters to estimate, and lists the model components created and used for dosing.
| Dosing Type | Description | SimBiology Model Components Used | Default Parameters to Estimate |
|---|---|---|---|
''(empty character vector) | No dose | The species ( | None |
SimBiology Model Builder app —
Command line —
| Assumes that the drug amount is increased instantly at the dose time. In the SimBiology model, the initial concentration of the drug is based on dose amount and volume of the compartment containing the drug. | The species ( | None |
SimBiology Model Builder app —
Command line —
| Assumes that the infused drug amount increases at a constant known absorption (or infusion) rate over a known duration. The imported data set must contain the rate and not an infusion duration. SimBiology uses this information to change the species concentration at the constant rate over the duration specified in the data set. | The species ( | None |
SimBiology Model Builder app —
Command line —
| Assumes that the drug is added at a constant rate over fixed, but unknown duration. |
|
|
SimBiology Model Builder app —
Command line —
| Assumes that the rate at which the drug is absorbed is not constant. In the SimBiology model, absorption rate is assumed to be governed by mass-action kinetics. |
|
|
Elimination Types
| Elimination Type | Description | SimBiology Model Components Created | Default Parameters to Estimate |
|---|---|---|---|
SimBiology Model Builder app — Command line —
| Assumes simple mass-action kinetics in the elimination of the drug. In the
SimBiology model, elimination is specified by mass-action kinetics with the
elimination rate constant specified by the forward rate parameter
(ke). |
|
|
SimBiology Model Builder app — Command line —
| Assumes simple mass-action kinetics in the elimination of the drug. In the
SimBiology model, similar to Linear {Elimination Rate,
Volume}. But, in addition, this option lets you specify the model in
terms of clearance (Cl) where, Cl = ke *
volume). |
|
|
SimBiology Model Builder app — Command line —
| Assumes that elimination is governed by Michaelis-Menten kinetics. |
|
|
Intercompartmental Clearance
The compartments created when you generate a SimBiology model form a chain and each pair of linked compartments are connected by a
transport reaction similar to linear elimination. The addition of two compartments,
C1 and C2, generates a reversible mass-action
reaction C1.Drug_C1 <-> C2.Drug. The forward rate parameter is the
compartmental clearance, Q12, divided by the
volume of C1. The reverse rate parameter is
Q12, divided by the volume of
C2.
The process of adding each pair of compartments in the chain
Cm and
Cn generates the following model
components:
A parameter
Qmnrepresenting the compartmental clearance between those two compartments. This parameter is added to the list of parameters to be estimated (Estimatedproperty ofPKModelMapobject).A parameter (
kmn) representing the rate of transfer of the drug fromCmtoCn, wherekmn = Qmn/Vm.A parameter (
knm) representing the rate ofCntoCm, whereknm = Qmn/Vn.A reversible mass-action reaction between the two compartments,
Cm.Drug_Cm <-> Cn.Drug_Cn, with forward rate parameterkmn, and reverse rate parameterknm.An initial assignment rule that initializes the value of the parameter
kmn, based on the initial values forCmandQmn.An initial assignment rule that initializes the value of the parameter
knm, based on the initial values forCnandQmn.
Unit Conversion for Imported Data
Unit conversion converts the matching physical quantities to one consistent unit system in order to resolve them. This conversion is in preparation for correct simulation, but SimBiology returns the physical quantities in the model in units that you specify.
Regardless of whether unit conversion is on or
off, you must express dosing data in amount. By default, Unit
Conversion is off, so you must ensure that units for the
data and the model are consistent with one another. If Unit Conversion
is on, you must specify units.
Parameters in the model have default units. If unit conversion is on,
you can change the units as long as the dimensions are consistent. These default units,
which you might use to specify the values for the initial guess, are as follows.
| Physical Quantity or Model Parameter | Unit |
|---|---|
| Central or peripheral compartment volume (Central or Peripheral) | liter |
| First-order elimination rate (ke) | 1/second |
| Michaelis constant (Km) | milligram/liter |
| Maximum reaction-velocity, Michaelis-Menten kinetics (Vm) | milligram/second |
| Clearance (Cl) | liter/second |
| Absorption duration (Tk0) | second |
| Absorption rate (ka) | 1/second |
Use the configuration settings options to turn unit conversion on or
off. For details, see Model Simulation.
For details on dimensional analysis for reaction rates, see How Reaction Rates Are Evaluated.
Create a Pharmacokinetic Model Using the Command Line
To create a PK model with the specified number of compartments, dosing type, and method of elimination:
Create a
PKModelDesignobject. ThePKModelDesignobject lets you specify the number of compartments, route of administration, and method of elimination, which SimBiology uses to construct the model object with the necessary compartments, species, reactions, and rules.pkm = PKModelDesign;
Add a compartment specifying the compartment name, and optionally, the type of dosing, and the method of elimination. Also specify whether the data contains a response variable measured in this compartment and whether the dose(s) have time lags. For example, specify a compartment named
Central, withBolusfor theDosingTypeproperty,linear-clearancefor theEliminationTypeproperty, andtruefor theHasResponseVariableproperty.pkc1 = addCompartment(pkm, 'Central', 'DosingType', 'Bolus', ... 'EliminationType', 'linear-clearance', ... 'HasResponseVariable', true);For a description of other
DosingTypeandEliminationTypeproperty values, see Dosing Types and Elimination Types.For a description of the
HasResponseVariableproperty, seeHasResponseVariable. At least one compartment in a model must have a response. Although SimBiology supports multiple responses per compartment, when adding compartments to aPKModelDesignobject, you are limited to one response per compartment.Note
To add a compartment that has a time lag associated with any dose that targets it, set the
HasLagproperty totrue:pkc_lag = addCompartment(pkm, 'Central', 'DosingType', 'Bolus', ... 'EliminationType', 'linear-clearance', ... 'HasResponseVariable', true, 'HasLag', true);Or after adding a compartment, set its
HasLagproperty totrue:pkc1.HasLag = true;
Optionally, add a second compartment named
Peripheral, with no dosing, no elimination, and no time lag. Set theHasResponseVariableproperty totrue. If you are using the tobramycin data set [1], skip this step and use only one compartment.pkc2 = addCompartment(pkm, 'Peripheral', 'HasResponseVariable', true);
The model construction process adds the necessary parameters, including a parameter representing intercompartmental clearance
Q. You can add more compartments by repeating this step. The addition of each compartment creates a chain of compartments in the order of compartment addition, with a bidirectional flow of the drug between compartments in the model.Use the handle to the compartment (
pkc1orpkc2), to change compartment properties.Construct a SimBiology model object.
[modelObj, PKModelMapObj] = pkm.construct
The
constructmethod returns a SimBiology model object (modelObj) and aPKModelMapobject (PKModelMapObj) that contains the mapping of the model components to the elements of the regression function.Note
If you change the
PKModelDesignobject, you must create a new model object using theconstructmethod. Changes to thePKModelDesigndo not automatically propagate to a previously constructed model object.Perform parameter fitting using Nonlinear Regression or Nonlinear Mixed-Effects Modeling.
The model object and the PKModelMap object are input
arguments for the sbionlmefit, sbionlmefitsa and
sbionlinfit functions used in parameter fitting.
See Also
Topics
- Calculate NCA Parameters and Fit Model to PK/PD Data Using SimBiology Model Analyzer
- Fit One-Compartment Model to Individual PK Profile
- Fit Two-Compartment Model to PK Profiles of Multiple Individuals
- Estimate Category-Specific PK Parameters for Multiple Individuals
- Estimate the Bioavailability of a Drug
- Model the Population Pharmacokinetics of Phenobarbital in Neonates