sbiofitmixed
Fit nonlinear mixed-effects model (requires Statistics and Machine Learning Toolbox software)
Syntax
Description
fitResults = sbiofitmixed(sm,grpData,ResponseMap,covEstiminfo)sm and returns a
NLMEResults object fitResults.
grpData is a groupedData
object specifying the data to fit.
ResponseMap defines the mapping between the model
components and response data in grpData.
covEstiminfo is a CovariateModel object or an array
of estimatedInfo objects that defines the parameters to be
estimated.
If the model contains active doses and variants, they are applied during the simulation.
fitResults = sbiofitmixed(sm,grpData,ResponseMap,covEstiminfo,dosing)dosing instead of using the active doses of the model
sm if there are any.
fitResults = sbiofitmixed(sm,grpData,ResponseMap,covEstiminfo,dosing,functionName)functionName that
must be either 'nlmefit' or
'nlmefitsa'.
fitResults = sbiofitmixed(sm,grpData,ResponseMap,covEstiminfo,dosing,functionName,opt)opt for the
estimation function functionName.
fitResults = sbiofitmixed(sm,grpData,ResponseMap,covEstiminfo,dosing,functionName,opt,variants)variants instead of
using any active variants of the model.
fitResults = sbiofitmixed(___,Name,Value)
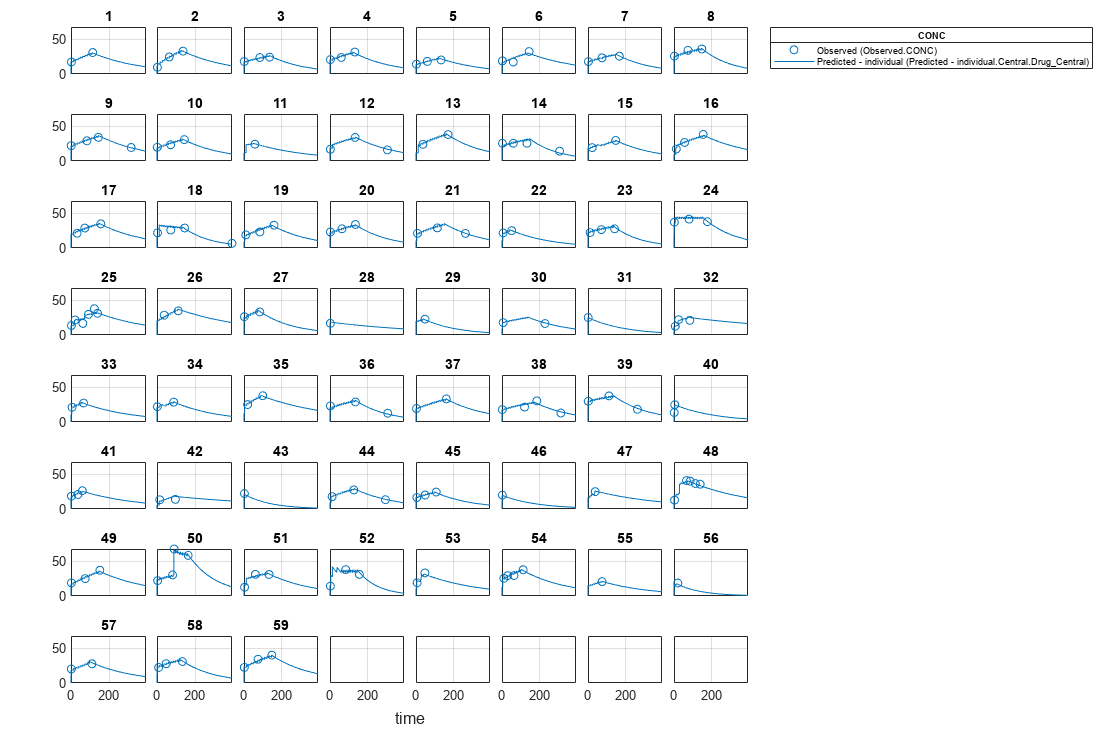
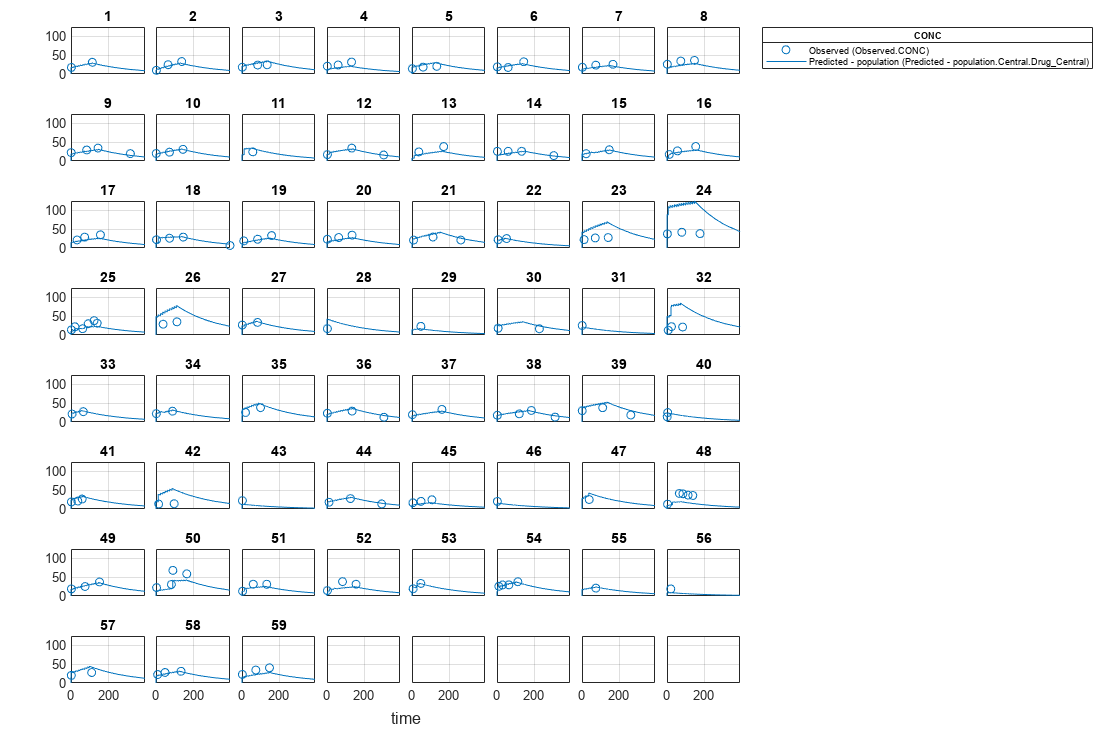
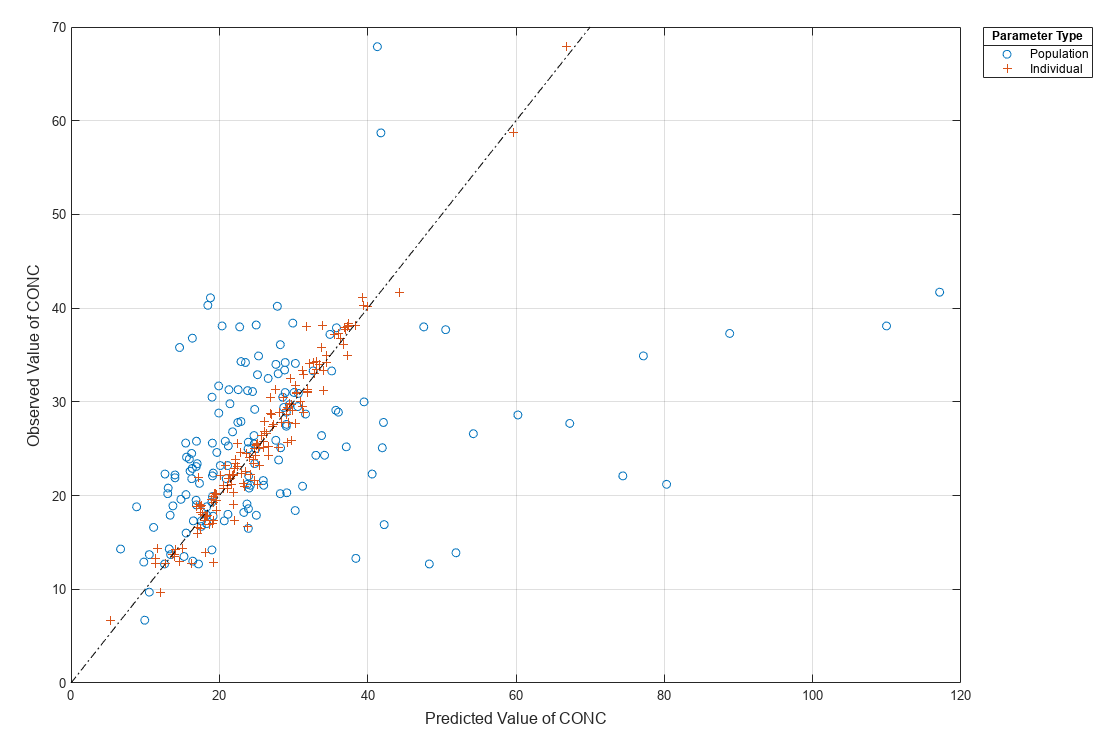
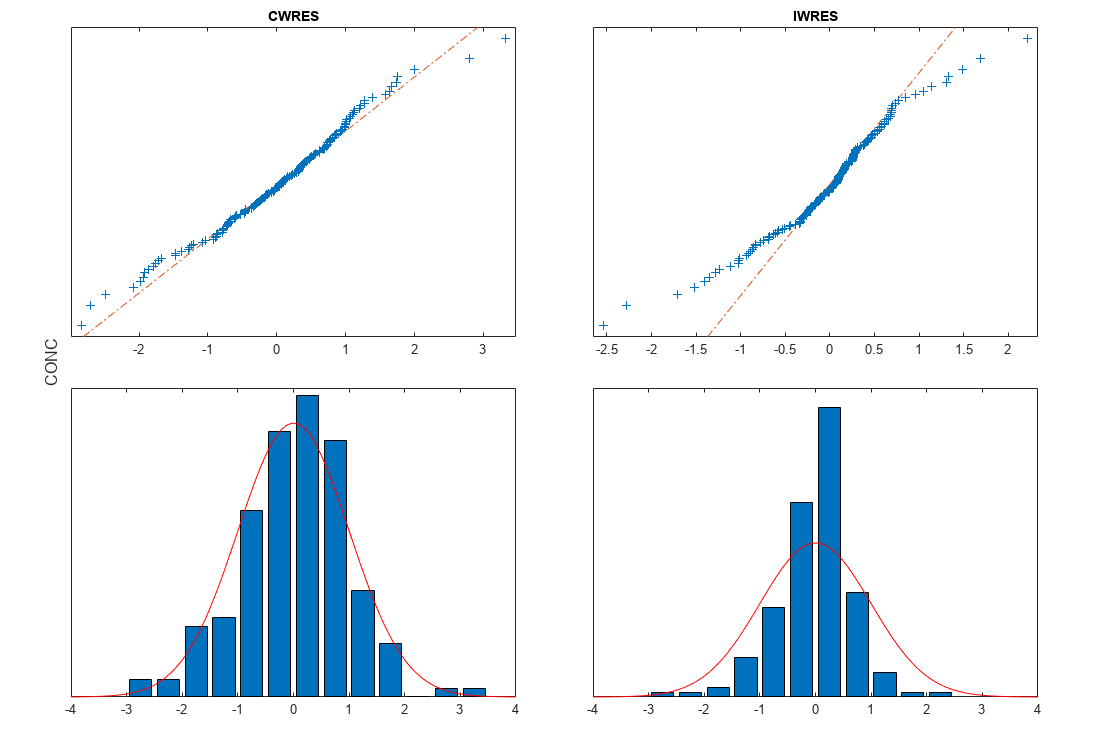
[ returns a vector of results objects fitResults,simDataI,simDataP]
= sbiofitmixed(_)fitResults,
vector of simulation results simDataI using individual-specific
parameter estimates, and vector of simulation results simDataP using
population parameter estimates.
Note
sbiofitmixedunifiessbionlmefitandsbionlmefitsaestimation functions. Usesbiofitmixedto perform nonlinear mixed-effects modeling and estimation.sbiofitmixedsimulates the model using aSimFunction object, which automatically accelerates simulations by default. Hence it is not necessary to runsbioacceleratebefore you callsbiofitmixed.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Grasela Jr, T.H., Donn, S.M. (1985) Neonatal population pharmacokinetics of phenobarbital derived from routine clinical data. Dev Pharmacol Ther. 8(6), 374–83.
Extended Capabilities
Version History
Introduced in R2014a
See Also
sbiofit | nlmefit (Statistics and Machine Learning Toolbox) | nlmefitsa (Statistics and Machine Learning Toolbox) | groupedData | EstimatedInfo object | NLMEResults object | sbiofitstatusplot | CovariateModel
Topics
- Model the Population Pharmacokinetics of Phenobarbital in Neonates
- What Is a Nonlinear Mixed-Effects Model?
- Nonlinear Mixed-Effects Modeling Workflow
- Specify a Covariate Model
- Specify an Error Model
- Maximum Likelihood Estimation
- Obtain the Fitting Status
- Supported Methods for Parameter Estimation in SimBiology
- Progress Plot
- Supported Files and Data Types
- Create Data File with SimBiology Definitions