selectbyname
Select simulation data by name from SimData object
Syntax
Description
[
returns the simulation time points t,x,names] = selectbyname(simdata,selectNames)t, the simulation data
x, and corresponding names for the states
specified by selectNames.
sdOut = selectbyname(simdata,selectNames)selectNames
as a SimData object sdOut.
___ = selectbyname(
returns the simulation data in the specified data format.simdata,selectNames,'Format',formatValue)
Examples
Load the model of glucose-insulin response. For details about the model, see the Background section in Simulate the Glucose-Insulin Response.
sbioloadproject('insulindemo.sbproj','m1');
Suppress an information warning that is issued during simulations.
warnSettings = warning('off', 'SimBiology:DimAnalysisNotDone_MatlabFcn_Dimensionless');
Simulate a single meal for a normal subject for 7 hours.
singleMeal = sbioselect(m1,'Name','Single Meal'); cs = getconfigset(m1,'active'); cs.StopTime = 7; sd = sbiosimulate(m1,singleMeal)
SimBiology Simulation Data
ModelName: Cobelli's Glucose-Insulin System
Logged Data:
Species: 15
Compartment: 0
Parameter: 24
Sensitivity: 0
Observable: 0
sbioplot(sd);
Select all species data logged in the SimData object sd.
[t,x,names] = select(sd,{'Type','species'});
namesnames = 15×1 cell
{'Glucose appearance.Dose' }
{'Glucose appearance.Stomach Glu Solid' }
{'Glucose appearance.Stomach Glu Tritur'}
{'Glucose appearance.Stomach Glu' }
{'Glucose appearance.Gut Glu' }
{'Glucose appearance.Plasma Glu' }
{'Glucose appearance.Plasma Glu Conc' }
{'Glucose appearance.Tissue Glu' }
{'Insulin secretion.Interstitial Ins' }
{'Insulin secretion.Portal Ins' }
{'Insulin secretion.Liver Ins' }
{'Insulin secretion.Plasma Ins' }
{'Insulin secretion.Plasma Ins Conc' }
{'Insulin secretion.Ins Delay 1' }
{'Insulin secretion.Ins Delay 2' }
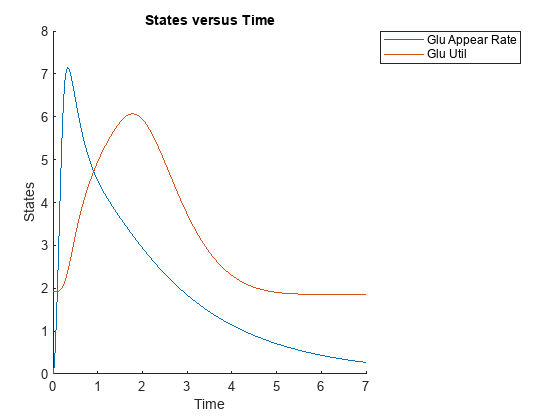
Plot data for the glucose rate of appearance and glucose utilization, namely Glu Appear Rate and Glu Util.
newsd = select(sd,{'Type','parameter','name',{'Glu Appear Rate'; 'Glu Util'}})
SimBiology Simulation Data
ModelName: Cobelli's Glucose-Insulin System
Logged Data:
Species: 0
Compartment: 0
Parameter: 2
Sensitivity: 0
Observable: 0
sbioplot(newsd);
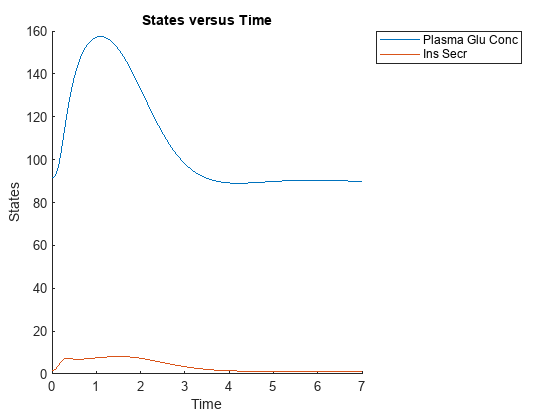
Compare data for the plasma glucose concentration (the species named Plasma Glu Conc) and insulin secretion rate (the parameter named Ins Secr). Use selectbyname to extract data by specifying the corresponding names.
newsd2 = selectbyname(sd,{'Plasma Glu Conc','Ins Secr'})
SimBiology Simulation Data
ModelName: Cobelli's Glucose-Insulin System
Logged Data:
Species: 1
Compartment: 0
Parameter: 1
Sensitivity: 0
Observable: 0
sbioplot(newsd2);
Select data for all species and parameters that have Glu in their names.
newsd3 = select(sd,{'Where','Name','regexp','Glu'})
SimBiology Simulation Data
ModelName: Cobelli's Glucose-Insulin System
Logged Data:
Species: 7
Compartment: 0
Parameter: 11
Sensitivity: 0
Observable: 0
newsd3.DataNames
ans = 18×1 cell
{'Stomach Glu Solid' }
{'Stomach Glu Tritur' }
{'Stomach Glu' }
{'Gut Glu' }
{'Plasma Glu' }
{'Plasma Glu Conc' }
{'Tissue Glu' }
{'Stomach Glu After Dosing'}
{'Glu Appear Rate' }
{'Glu Prod' }
{'Plasma Glu Conc Rate' }
{'Ins Dep Glu Util' }
{'Glu Util' }
{'Glu Excretion' }
{'Glu Excretion Mode' }
{'Delayed Glu Signal' }
{'Delayed Glu Signal Mode' }
{'Basal Glu Prod' }
You can also return the selected data as a structure.
sdStruct = select(sd,{'Where','Name','regexp','Glu'},'Format','struct');Restore the warning settings.
warning(warnSettings);
Input Arguments
Simulation data, specified as a SimData object or array of
SimData objects.
Names of states that you want to select data for, specified as a character vector, string, string vector, or cell array of character vectors.
Example: {'x1','x2','x3'}
Data Types: char | string | cell
Simulation data format, specified as a character vector or string. Some formats require you to specify only one output argument. The valid formats follow.
'num'— This format returns simulation time points and simulation data in numeric arrays and the names of quantities and sensitivities as a cell array. This format is the default when you rungetdatawith multiple output arguments.'nummetadata'— This format returns a cell array of metadata structures instead of the names of quantities and sensitivities as the third output argument.'numqualnames'— This format returns qualified names in the third output argument to resolve ambiguities.
You must specify only one output argument for the following formats.
'simdata'— This format returns data in a newSimDataobject or an array ofSimDataobjects. This format is the default when you specify a single output argument.'struct'— This format returns a structure or structure array that contains both data and metadata.'ts'— This format returns data as a cell array.If
simdatais scalar, the cell array is an m-by-1 array, where each element is atimeseriesobject. m is the number of quantities and sensitivities logged during the simulation.If
simdatais not scalar, the cell array is k-by-1, where each element of the cell array is an m-by-1 cell array oftimeseriesobjects. k is the size ofsimdata, and m is the number of quantities or sensitivities in eachSimDataobject insimdata. In other words, the function returns an individual time series for each state or column and for eachSimDataobject insimdata.
'tslumped'— This format returns the data as a cell array oftimeseriesobjects, combining data from eachSimDataobject into a single time series.
Output Arguments
Simulation time points, returned as a numeric vector or cell array. If
simdata is scalar, t is an
n-by-1 vector, where n is the number of time
points. If simdata is an array of objects, t
is a k-by-1 cell array, where k is the size of
simdata.
Simulation data, returned as a numeric matrix or cell array. If simdata
is scalar, x is an n-by-m
matrix, where n is the number of time points and m
is the number of quantities and sensitivities logged during the simulation. If
simdata is an array of objects, x is a
k-by-1 cell array, where k is the size of
simdata.
Names of quantities and sensitivities logged during the simulation, returned as a cell array.
If simdata is scalar, names is an
m-by-1 cell array. If simdata is an array of
objects, names is a k-by-1 cell array, where
k is the size of simdata.
Simulation results, returned as a SimData object.
Version History
Introduced in R2007b
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)