edge
Description
e = edge(Mdl,Tbl,ResponseVarName)e)
for the generalized additive model Mdl using the predictor data in
Tbl and the true class labels in
Tbl.ResponseVarName.
e = edge(___,Name,Value)
Examples
Estimate the test sample classification margins and edge of a generalized additive model. The test sample margins are the observed true class scores minus the false class scores, and the test sample edge is the mean of the margins.
Load the fisheriris data set. Create X as a numeric matrix that contains two sepal and two petal measurements for versicolor and virginica irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris inds = strcmp(species,'versicolor') | strcmp(species,'virginica'); X = meas(inds,:); Y = species(inds,:);
Randomly partition observations into a training set and a test set with stratification, using the class information in Y. Specify a 30% holdout sample for testing.
rng('default') % For reproducibility cv = cvpartition(Y,'HoldOut',0.30);
Extract the training and test indices.
trainInds = training(cv); testInds = test(cv);
Specify the training and test data sets.
XTrain = X(trainInds,:); YTrain = Y(trainInds); XTest = X(testInds,:); YTest = Y(testInds);
Train a GAM using the predictors XTrain and class labels YTrain. A recommended practice is to specify the class names.
Mdl = fitcgam(XTrain,YTrain,'ClassNames',{'versicolor','virginica'});
Mdl is a ClassificationGAM model object.
Estimate the test sample classification margins and edge.
m = margin(Mdl,XTest,YTest); e = edge(Mdl,XTest,YTest)
e = 0.8000
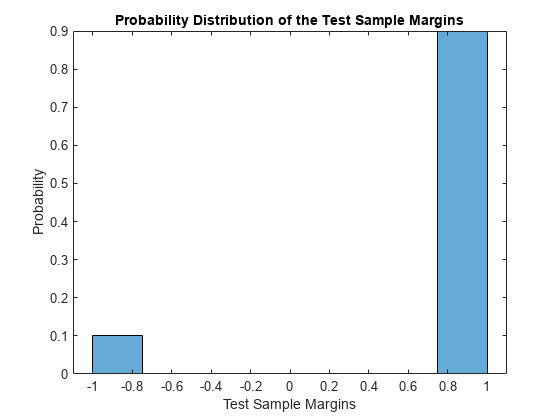
Display the histogram of the test sample classification margins.
histogram(m,length(unique(m)),'Normalization','probability') xlabel('Test Sample Margins') ylabel('Probability') title('Probability Distribution of the Test Sample Margins')
Estimate the test sample weighted edge (the weighted average of margins) of a generalized additive model.
Load the fisheriris data set. Create X as a numeric matrix that contains two sepal and two petal measurements for versicolor and virginica irises. Create Y as a cell array of character vectors that contains the corresponding iris species.
load fisheriris idx1 = strcmp(species,'versicolor') | strcmp(species,'virginica'); X = meas(idx1,:); Y = species(idx1,:);
Suppose that the quality of some measurements is lower because they were measured with older technology. To simulate this effect, add noise to a random subset of 20 measurements.
rng('default') % For reproducibility idx2 = randperm(size(X,1),20); X(idx2,:) = X(idx2,:) + 2*randn(20,size(X,2));
Randomly partition observations into a training set and a test set with stratification, using the class information in Y. Specify a 30% holdout sample for testing.
cv = cvpartition(Y,'HoldOut',0.30);Extract the training and test indices.
trainInds = training(cv); testInds = test(cv);
Specify the training and test data sets.
XTrain = X(trainInds,:); YTrain = Y(trainInds); XTest = X(testInds,:); YTest = Y(testInds);
Train a GAM using the predictors XTrain and class labels YTrain. A recommended practice is to specify the class names.
Mdl = fitcgam(XTrain,YTrain,'ClassNames',{'versicolor','virginica'});
Mdl is a ClassificationGAM model object.
Estimate the test sample edge.
e = edge(Mdl,XTest,YTest)
e = 0.8000
The average margin is approximately 0.80.
One way to reduce the effect of the noisy measurements is to assign them less weight than the other observations. Define a weight vector that gives the higher quality observations twice the weight of the other observations.
n = size(X,1); weights = ones(size(X,1),1); weights(idx2) = 0.5; weightsTrain = weights(trainInds); weightsTest = weights(testInds);
Train a GAM using the predictors XTrain, class labels YTrain, and weights weightsTrain.
Mdl_W = fitcgam(XTrain,YTrain,'Weights',weightsTrain,... 'ClassNames',{'versicolor','virginica'});
Estimate the test sample weighted edge using the weighting scheme.
e_W = edge(Mdl_W,XTest,YTest,'Weights',weightsTest)e_W = 0.8770
The weighted average margin is approximately 0.88. This result indicates that, on average, the labels from weighted classifier labels have higher confidence.
Compare a GAM with linear terms to a GAM with both linear and interaction terms by examining the test sample margins and edge. Based solely on this comparison, the classifier with the highest margins and edge is the best model.
Load the ionosphere data set. This data set has 34 predictors and 351 binary responses for radar returns, either bad ('b') or good ('g').
load ionosphereRandomly partition observations into a training set and a test set with stratification, using the class information in Y. Specify a 30% holdout sample for testing.
rng('default') % For reproducibility cv = cvpartition(Y,'Holdout',0.30);
Extract the training and test indices.
trainInds = training(cv); testInds = test(cv);
Specify the training and test data sets.
XTrain = X(trainInds,:); YTrain = Y(trainInds); XTest = X(testInds,:); YTest = Y(testInds);
Train a GAM that contains both linear and interaction terms for predictors. Specify to include all available interaction terms whose p-values are not greater than 0.05.
Mdl = fitcgam(XTrain,YTrain,'Interactions','all','MaxPValue',0.05)
Mdl =
ClassificationGAM
ResponseName: 'Y'
CategoricalPredictors: []
ClassNames: {'b' 'g'}
ScoreTransform: 'logit'
Intercept: 3.0398
Interactions: [561×2 double]
NumObservations: 246
Properties, Methods
Mdl is a ClassificationGAM model object. Mdl includes all available interaction terms.
Estimate the test sample margins and edge for Mdl.
M = margin(Mdl,XTest,YTest); E = edge(Mdl,XTest,YTest)
E = 0.7848
Estimate the test sample margins and edge for Mdl without including interaction terms.
M_nointeractions = margin(Mdl,XTest,YTest,'IncludeInteractions',false); E_nointeractions = edge(Mdl,XTest,YTest,'IncludeInteractions',false)
E_nointeractions = 0.7871
Display the distributions of the margins using box plots.
boxplot([M M_nointeractions],'Labels',{'Linear and Interaction Terms','Linear Terms Only'}) title('Box Plots of Test Sample Margins')
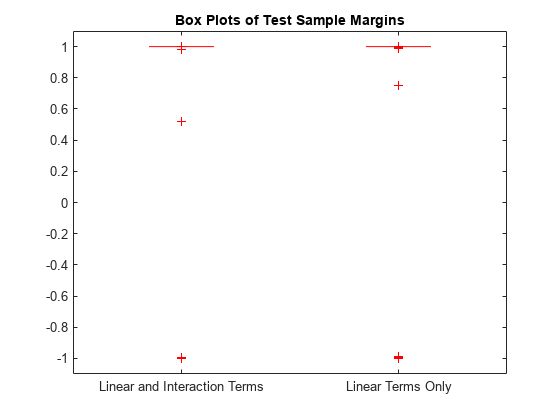
The margins M and M_nointeractions have a similar distribution, but the test sample edge of the classifier with only linear terms is larger. Classifiers that yield relatively large margins are preferred.
Input Arguments
Generalized additive model, specified as a ClassificationGAM or CompactClassificationGAM model object.
Sample data, specified as a table. Each row of Tbl corresponds to one observation, and each column corresponds to one predictor variable. Multicolumn variables and cell arrays other than cell arrays of character vectors are not allowed.
Tbl must contain all the predictors used to train
Mdl. Optionally, Tbl can contain a column
for the response variable and a column for the observation weights.
The response variable must have the same data type as
Mdl.Y. (The software treats string arrays as cell arrays of character vectors.) If the response variable inTblhas the same name as the response variable used to trainMdl, then you do not need to specifyResponseVarName.The weight values must be a numeric vector. You must specify the observation weights in
Tblby using'Weights'.
If you trained Mdl using sample data contained in a table, then the input data for edge must also be in a table.
Data Types: table
Response variable name, specified as a character vector or string scalar containing the name
of the response variable in Tbl. For example, if the response
variable Y is stored in Tbl.Y, then specify it as
'Y'.
Data Types: char | string
Class labels, specified as a categorical, character, or string array, a logical or
numeric vector, or a cell array of character vectors. Each row of Y
represents the classification of the corresponding row of X or
Tbl.
Y must have the same data type as Mdl.Y. (The software treats string arrays as cell arrays of character
vectors.)
Data Types: single | double | categorical | logical | char | string | cell
Predictor data, specified as a numeric matrix. Each row of X corresponds to one observation, and each column corresponds to one predictor variable.
If you trained Mdl using sample data contained in a matrix, then the input data for edge must also be in a matrix.
Data Types: single | double
Name-Value Arguments
Specify optional pairs of arguments as
Name1=Value1,...,NameN=ValueN, where Name is
the argument name and Value is the corresponding value.
Name-value arguments must appear after other arguments, but the order of the
pairs does not matter.
Before R2021a, use commas to separate each name and value, and enclose
Name in quotes.
Example: 'IncludeInteractions',false,'Weights',w specifies to exclude
interaction terms from the model and to use the observation weights
w.
Flag to include interaction terms of the model, specified as true or
false.
The default 'IncludeInteractions' value is true if Mdl contains interaction terms. The value must be false if the model does not contain interaction terms.
Example: 'IncludeInteractions',false
Data Types: logical
Observation weights, specified as a vector of scalar values or the name of a variable in Tbl. The software weights the observations in each row of X or Tbl with the corresponding value in Weights. The size of Weights must equal the number of rows in X or Tbl.
If you specify the input data as a table Tbl, then
Weights can be the name of a variable in Tbl
that contains a numeric vector. In this case, you must specify
Weights as a character vector or string scalar. For example, if
the weights vector W is stored in Tbl.W, then
specify it as 'W'.
edge normalizes the weights in each class to add up to the value of the prior probability of the respective class.
Data Types: single | double | char | string
More About
The classification edge is the weighted mean of the classification margins.
One way to choose among multiple classifiers, for example to perform feature selection, is to choose the classifier that yields the greatest edge.
The classification margin for binary classification is, for each observation, the difference between the classification score for the true class and the classification score for the false class.
If the margins are on the same scale (that is, the score values are based on the same score transformation), then they serve as a classification confidence measure. Among multiple classifiers, those that yield greater margins are better.
Version History
Introduced in R2021a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)