Generate Code at Command Line Using Model Exported from Machine Learning App
The Classification Learner and Regression Learner apps are well suited for choosing and training machine learning models interactively, and offer several ways to generate code. After training a model in an app, you can use the Generate Function button in the Export section to generate MATLAB code for training the model with new data at the command line. The Export Model to Coder option, available from the Export Model button, exports a trained model from the app to MATLAB® Coder™ so you can generate C/C++ code for the model.
The apps do not support all model types for export to MATLAB coder.
Also, in some cases, you might want to customize code generation options. In these
situations, you need to generate C/C++ code for a trained app model at the MATLAB command
line using the saveLearnerForCoder, loadLearnerForCoder, and codegen (MATLAB Coder) functions.
This example shows how to generate C code from a function that predicts labels by using a classification model exported from Classification Learner. The process is the same for working with a model exported from Regression Learner. The example builds a model that predicts the credit rating of a business given various financial ratios, according to these steps:
Use the credit rating data set in the file
CreditRating_Historical.dat, which is included with Statistics and Machine Learning Toolbox™.Reduce the data dimensionality using principal component analysis (PCA).
Train a set of models that support code generation for label prediction.
Export the model with the minimum 5-fold, cross-validated classification accuracy.
Generate C code from an entry-point function that transforms the new predictor data and then predicts corresponding labels using the exported model.
Load Sample Data
Load sample data and import the data into the Classification Learner app. Review the data using scatter plots and remove unnecessary predictors.
Use readtable to load the historical credit rating data set in
the file CreditRating_Historical.dat into a table.
creditrating = readtable('CreditRating_Historical.dat');
On the Apps tab, click Classification Learner.
In Classification Learner, on the Classification Learner tab, in the File section, click New Session and select From Workspace.
In the New Session from Workspace dialog box, select the table
creditrating. All variables, except the one identified as the
response, are double-precision numeric vectors. Click Start
Session to compare classification models based on the 5-fold,
cross-validated classification accuracy.
Classification Learner loads the data and plots a scatter plot of the variables
WC_TA versus ID. Because identification numbers
are not helpful to display in a plot, choose RE_TA for
X under Predictors.
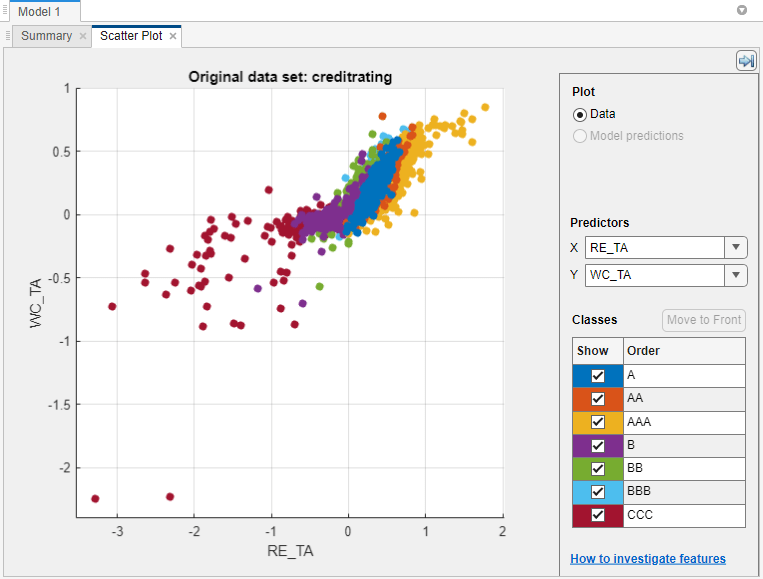
The scatter plot suggests that the two variables can separate the classes
AAA, BBB, BB, and
CCC fairly well. However, the observations corresponding to the
remaining classes are mixed into these classes.
Identification numbers are not helpful for prediction. Therefore, in the Options section of the Classification Learner tab, click Feature Selection. In the Default Feature Selection tab, clear the ID check box, and click Save and Apply. You can also remove unnecessary predictors from the beginning by using the check boxes in the New Session from Workspace dialog box. This example shows how to remove unused predictors for code generation when you have included all predictors.
Enable PCA
Enable PCA to reduce the data dimensionality.
In the Options section of the Classification Learner tab, click PCA. In the Default PCA Options dialog box, select Enable PCA, and click Save and Apply. This action applies PCA to the predictor data, and then transforms the data before training the models. Classification Learner uses only components that collectively explain 95% of the variability.
Train Models
Train a set of models that support code generation for label prediction. For a list of models in Classification Learner that support code generation, see Export Classification Model to MATLAB Coder to Generate C/C++ Code.
Select the following classification models and options, which support code generation for label prediction, and then perform cross-validation (for more details, see Introduction to Code Generation). To select each model, in the Models section, click the Show more arrow, and then click the model.
| Models and Options to Select | Description |
|---|---|
| Under Decision Trees, select All Trees | Classification trees of various complexities |
| Under Support Vector Machines, select All SVMs | SVMs of various complexities and using various kernels. Complex SVMs require time to fit. |
| Under Ensemble Classifiers, select Boosted Trees. In the model Summary tab, under Model Hyperparameters, reduce Maximum number of splits to 5 and increase Number of learners to 100. | Boosted ensemble of classification trees |
| Under Ensemble Classifiers, select Bagged Trees. In the model Summary tab, under Model Hyperparameters, reduce Maximum number of splits to 50 and increase Number of learners to 100. | Random forest of classification trees |
After selecting the models and specifying any options, delete the default fine tree model (model 1). Right-click the model in the Models pane and select Delete. Then, in the Train section, click Train All and select Train All.
After the app cross-validates each model type, the Models pane displays each model and its 5-fold, cross-validated classification accuracy. The app Accuracy (Validation) value of the model with the highest accuracy is highlighted with a box.
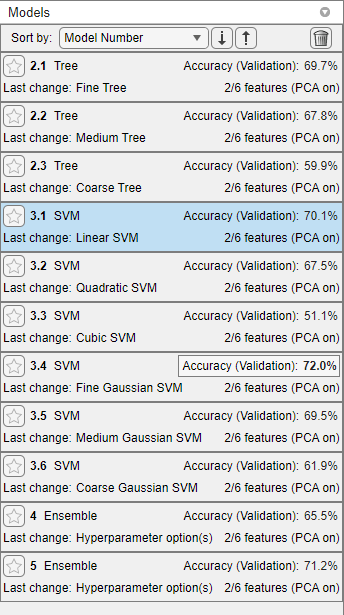
Select the model that has the highest 5-fold, cross-validated classification accuracy, which is the error-correcting output codes (ECOC) model of Fine Gaussian SVM learners. With PCA enabled, Classification Learner uses two predictors out of six.
In the Plot and Interpret section, click the arrow to open the gallery, and then click Confusion Matrix (Validation) in the Validation Results group.
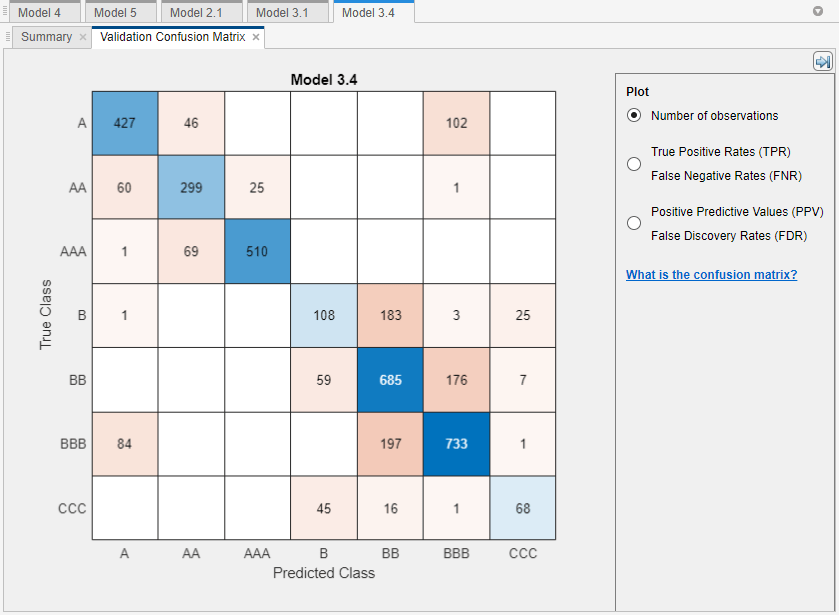
The model does well distinguishing between A,
B, and C classes. However, the model does not do
as well distinguishing between particular levels within those groups, the lower B levels
in particular.
Export Model to Workspace
Export the model to the MATLAB Workspace and save the model using saveLearnerForCoder.
On the Classification Learner tab, click Export, click Export Model, and select Export Model. In the Export Classification Model dialog box, clear the check box to exclude the training data from the exported model. Click OK to export the compact model.
The structure trainedModel appears in the MATLAB Workspace. The field ClassificationSVM of
trainedModel contains the compact model.
At the command line, save the compact model to a file called
ClassificationLearnerModel.mat in your current folder.
saveLearnerForCoder(trainedModel.ClassificationSVM,'ClassificationLearnerModel')Generate C Code for Prediction
Prediction using the object functions requires a trained model object, but the
-args option of codegen (MATLAB Coder) does not accept such objects. Work around this limitation by using
saveLearnerForCoder and loadLearnerForCoder. Save a trained model by using
saveLearnerForCoder. Then, define an entry-point function that loads
the saved model by using loadLearnerForCoder and calls the
predict function. Finally, use codegen to
generate code for the entry-point function.
Preprocess Data
Preprocess new data in the same way you preprocess the training data.
To preprocess, you need the following three model parameters:
removeVars— Column vector of at mostpelements identifying indices of variables to remove from the data, wherepis the number of predictor variables in the raw datapcaCenters— Row vector of exactlyqPCA centerspcaCoefficients—q-by-rmatrix of PCA coefficients, whereris at mostq
Specify the indices of predictor variables that you removed while selecting data
using Feature Selection in Classification Learner. Extract the
PCA statistics from trainedModel.
removeVars = 1; pcaCenters = trainedModel.PCACenters; pcaCoefficients = trainedModel.PCACoefficients;
Save the model parameters to a file named ModelParameters.mat
in your current folder.
save('ModelParameters.mat','removeVars','pcaCenters','pcaCoefficients');
Define Entry-Point Function
An entry-point function is a function you define for code
generation. Because you cannot call any function at the top level using
codegen, you must define an entry-point function that calls
code-generation-enabled functions, and then generate C/C++ code for the entry-point
function by using codegen.
In your current folder, define a function named mypredictCL.m that:
Accepts a numeric matrix (
X) of raw observations containing the same predictor variables as the ones passed into Classification LearnerLoads the classification model in
ClassificationLearnerModel.matand the model parameters inModelParameters.matRemoves the predictor variables corresponding to the indices in
removeVarsTransforms the remaining predictor data using the PCA centers (
pcaCenters) and coefficients (pcaCoefficients) estimated by Classification LearnerReturns predicted labels using the model
function label = mypredictCL(X) %#codegen %MYPREDICTCL Classify credit rating using model exported from %Classification Learner % MYPREDICTCL loads trained classification model (SVM) and model % parameters (removeVars, pcaCenters, and pcaCoefficients), removes the % columns of the raw matrix of predictor data in X corresponding to the % indices in removeVars, transforms the resulting matrix using the PCA % centers in pcaCenters and PCA coefficients in pcaCoefficients, and then % uses the transformed data to classify credit ratings. X is a numeric % matrix with n rows and 7 columns. label is an n-by-1 cell array of % predicted labels. % Load trained classification model and model parameters SVM = loadLearnerForCoder('ClassificationLearnerModel'); data = coder.load('ModelParameters'); removeVars = data.removeVars; pcaCenters = data.pcaCenters; pcaCoefficients = data.pcaCoefficients; SVM = loadLearnerForCoder('ClassificationLearnerModel'); % Remove unused predictor variables keepvars = 1:size(X,2); idx = ~ismember(keepvars,removeVars); keepvars = keepvars(idx); XwoID = X(:,keepvars); % Transform predictors via PCA Xpca = bsxfun(@minus,XwoID,pcaCenters)*pcaCoefficients; % Generate label from SVM label = predict(SVM,Xpca); end
Generate Code
Because C and C++ are statically typed languages, you must
determine the properties of all variables in the entry-point function at compile time.
Specify variable-size arguments using coder.typeof (MATLAB Coder) and generate code using the arguments.
Create a double-precision matrix called x for code generation
using coder.typeof (MATLAB Coder). Specify that the number of
rows of x is arbitrary, but that x must have
p columns.
p = size(creditrating,2) - 1; x = coder.typeof(0,[Inf,p],[1 0]);
For more details about specifying variable-size arguments, see Specify Variable-Size Arguments for Code Generation.
Generate a MEX function from mypredictCL.m. Use the
-args option to specify x as an
argument.
codegen mypredictCL -args x
codegen generates the MEX file
mypredictCL_mex.mexw64 in your current folder. The file extension
depends on your platform.
Verify Generated Code
Verify that the MEX function returns the expected labels.
Remove the response variable from the original data set, and then randomly draw 15 observations.
rng('default'); % For reproducibility m = 15; testsampleT = datasample(creditrating(:,1:(end - 1)),m);
Predict corresponding labels by using predictFcn in the
classification model trained by Classification Learner.
testLabels = trainedModel.predictFcn(testsampleT);
Convert the resulting table to a matrix.
testsample = table2array(testsampleT);
The columns of testsample correspond to the columns of the
predictor data loaded by Classification Learner.
Pass the test data to mypredictCL. The function
mypredictCL predicts corresponding labels by using
predict and the classification model trained by Classification
Learner.
testLabelsPredict = mypredictCL(testsample);
Predict corresponding labels by using the generated MEX function
mypredictCL_mex.
testLabelsMEX = mypredictCL_mex(testsample);
Compare the sets of predictions.
isequal(testLabels,testLabelsMEX,testLabelsPredict)
ans = logical 1
isequal returns logical 1 (true) if all
the inputs are equal. predictFcn, mypredictCL,
and the MEX function return the same values.
See Also
loadLearnerForCoder | saveLearnerForCoder | coder.typeof (MATLAB Coder) | codegen (MATLAB Coder) | learnerCoderConfigurer
Topics
- Classification Learner App
- Predict Responses Using RegressionSVM Predict Block
- Introduction to Code Generation
- Code Generation for Prediction of Machine Learning Model at Command Line
- Code Generation for Prediction of Machine Learning Model Using MATLAB Coder App
- Code Generation for Prediction and Update Using Coder Configurer
- Specify Variable-Size Arguments for Code Generation
- Apply PCA to New Data and Generate C/C++ Code