predict
Description
fairnessLabels = predict(thresholder,Tbl)fairnessThresholder object
thresholder using the table Tbl.
fairnessLabels = predict(thresholder,X,attribute)fairnessThresholder object
thresholder using the matrix data X and the
sensitive attribute specified by attribute.
Examples
Train a tree ensemble for binary classification, and compute the disparate impact for each group in the sensitive attribute. To reduce the disparate impact value of the nonreference group, adjust the score threshold for classifying observations.
Load the data census1994, which contains the data set adultdata and the test data set adulttest. The data sets consist of demographic information from the US Census Bureau that can be used to predict whether an individual makes over $50,000 per year. Preview the first few rows of adultdata.
load census1994
head(adultdata) age workClass fnlwgt education education_num marital_status occupation relationship race sex capital_gain capital_loss hours_per_week native_country salary
___ ________________ __________ _________ _____________ _____________________ _________________ _____________ _____ ______ ____________ ____________ ______________ ______________ ______
39 State-gov 77516 Bachelors 13 Never-married Adm-clerical Not-in-family White Male 2174 0 40 United-States <=50K
50 Self-emp-not-inc 83311 Bachelors 13 Married-civ-spouse Exec-managerial Husband White Male 0 0 13 United-States <=50K
38 Private 2.1565e+05 HS-grad 9 Divorced Handlers-cleaners Not-in-family White Male 0 0 40 United-States <=50K
53 Private 2.3472e+05 11th 7 Married-civ-spouse Handlers-cleaners Husband Black Male 0 0 40 United-States <=50K
28 Private 3.3841e+05 Bachelors 13 Married-civ-spouse Prof-specialty Wife Black Female 0 0 40 Cuba <=50K
37 Private 2.8458e+05 Masters 14 Married-civ-spouse Exec-managerial Wife White Female 0 0 40 United-States <=50K
49 Private 1.6019e+05 9th 5 Married-spouse-absent Other-service Not-in-family Black Female 0 0 16 Jamaica <=50K
52 Self-emp-not-inc 2.0964e+05 HS-grad 9 Married-civ-spouse Exec-managerial Husband White Male 0 0 45 United-States >50K
Each row contains the demographic information for one adult. The information includes sensitive attributes, such as age, marital_status, relationship, race, and sex. The third column flnwgt contains observation weights, and the last column salary shows whether a person has a salary less than or equal to $50,000 per year (<=50K) or greater than $50,000 per year (>50K).
Remove observations with missing values.
adultdata = rmmissing(adultdata); adulttest = rmmissing(adulttest);
Partition adultdata into training and validation sets. Use 60% of the observations for the training set trainingData and 40% of the observations for the validation set validationData.
rng("default") % For reproducibility c = cvpartition(adultdata.salary,"Holdout",0.4); trainingIdx = training(c); validationIdx = test(c); trainingData = adultdata(trainingIdx,:); validationData = adultdata(validationIdx,:);
Train a boosted ensemble of trees using the training data set trainingData. Specify the response variable, predictor variables, and observation weights by using the variable names in the adultdata table. Use random undersampling boosting as the ensemble aggregation method.
predictors = ["capital_gain","capital_loss","education", ... "education_num","hours_per_week","occupation","workClass"]; Mdl = fitcensemble(trainingData,"salary", ... PredictorNames=predictors, ... Weights="fnlwgt",Method="RUSBoost");
Predict salary values for the observations in the test data set adulttest, and calculate the classification error.
labels = predict(Mdl,adulttest); L = loss(Mdl,adulttest)
L = 0.2080
The model accurately predicts the salary categorization for approximately 80% of the test set observations.
Compute fairness metrics with respect to the sensitive attribute sex by using the test set model predictions. In particular, find the disparate impact for each group in sex. Use the report and plot object functions of fairnessMetrics to display the results.
metricsResults = fairnessMetrics(adulttest,"salary", ... SensitiveAttributeNames="sex",Predictions=labels, ... ModelNames="Ensemble",Weights="fnlwgt"); metricsResults.PositiveClass
ans = categorical
>50K
metricsResults.ReferenceGroup
ans = 'Male'
report(metricsResults,BiasMetrics="DisparateImpact")ans=2×4 table
ModelNames SensitiveAttributeNames Groups DisparateImpact
__________ _______________________ ______ _______________
Ensemble sex Female 0.73792
Ensemble sex Male 1
plot(metricsResults,"DisparateImpact")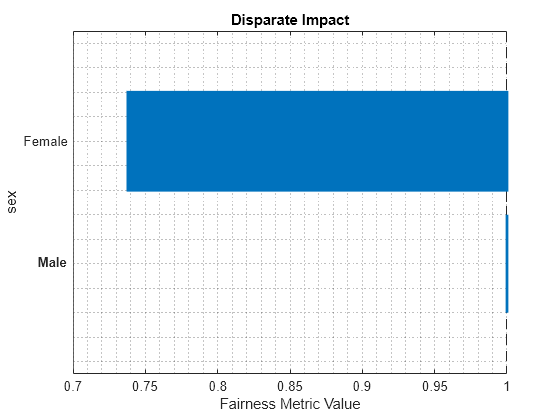
For the nonreference group (Female), the disparate impact value is the proportion of predictions in the group with a positive class value (>50K) divided by the proportion of predictions in the reference group (Male) with a positive class value. Ideally, disparate impact values are close to 1.
To try to improve the nonreference group disparate impact value, you can adjust model predictions by using the fairnessThresholder function. The function uses validation data to search for an optimal score threshold that maximizes accuracy while satisfying fairness bounds. For observations in the critical region below the optimal threshold, the function changes the labels so that the fairness constraints hold for the reference and nonreference groups. By default, the function tries to find a score threshold so that the disparate impact value for the nonreference group is in the range [0.8,1.25].
fairnessMdl = fairnessThresholder(Mdl,validationData,"sex","salary")
fairnessMdl =
fairnessThresholder with properties:
Learner: [1×1 classreg.learning.classif.CompactClassificationEnsemble]
SensitiveAttribute: 'sex'
ReferenceGroups: Male
ResponseName: 'salary'
PositiveClass: >50K
ScoreThreshold: 1.6749
BiasMetric: 'DisparateImpact'
BiasMetricValue: 0.9702
BiasMetricRange: [0.8000 1.2500]
ValidationLoss: 0.2017
fairnessMdl is a fairnessThresholder model object. Note that the predict function of the ensemble model Mdl returns scores that are not posterior probabilities. Scores are in the range instead, and the maximum score for each observation is greater than 0. For observations whose maximum scores are less than the new score threshold (fairnessMdl.ScoreThreshold), the predict function of the fairnessMdl object adjusts the prediction. If the observation is in the nonreference group, the function predicts the observation into the positive class. If the observation is in the reference group, the function predicts the observation into the negative class. These adjustments do not always result in a change in the predicted label.
Adjust the test set predictions by using the new score threshold, and calculate the classification error.
fairnessLabels = predict(fairnessMdl,adulttest); fairnessLoss = loss(fairnessMdl,adulttest)
fairnessLoss = 0.2064
The new classification error is similar to the original classification error.
Compare the disparate impact values across the two sets of test predictions: the original predictions computed using Mdl and the adjusted predictions computed using fairnessMdl.
newMetricsResults = fairnessMetrics(adulttest,"salary", ... SensitiveAttributeNames="sex",Predictions=[labels,fairnessLabels], ... ModelNames=["Original","Adjusted"],Weights="fnlwgt"); newMetricsResults.PositiveClass
ans = categorical
>50K
newMetricsResults.ReferenceGroup
ans = 'Male'
report(newMetricsResults,BiasMetrics="DisparateImpact")ans=2×5 table
Metrics SensitiveAttributeNames Groups Original Adjusted
_______________ _______________________ ______ ________ ________
DisparateImpact sex Female 0.73792 1.0048
DisparateImpact sex Male 1 1
plot(newMetricsResults,"di")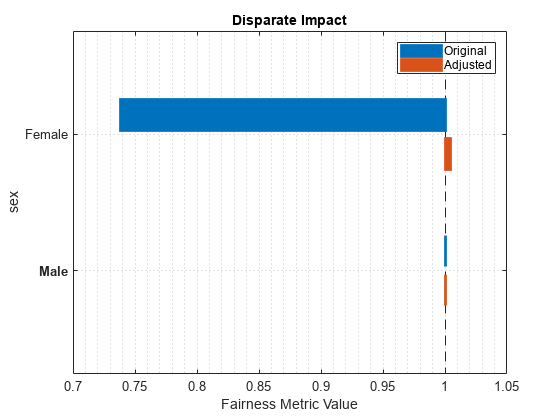
The disparate impact value for the nonreference group (Female) is closer to 1 when you use the adjusted predictions.
Train a support vector machine (SVM) model, and compute the statistical parity difference (SPD) for each group in the sensitive attribute. To reduce the SPD value of the nonreference group, adjust the score threshold for classifying observations.
Load the patients data set, which contains medical information for 100 patients. Convert the Gender and Smoker variables to categorical variables. Specify the descriptive category names Smoker and Nonsmoker rather than 1 and 0.
load patients Gender = categorical(Gender); Smoker = categorical(Smoker,logical([1 0]), ... ["Smoker","Nonsmoker"]);
Create a matrix containing the continuous predictors Diastolic and Systolic. Specify Gender as the sensitive attribute and Smoker as the response variable.
X = [Diastolic,Systolic]; attribute = Gender; Y = Smoker;
Partition the data into training and validation sets. Use half of the observations for training and half of the observations for validation.
rng("default") % For reproducibility cv = cvpartition(Y,"Holdout",0.5); trainX = X(training(cv),:); trainAttribute = attribute(training(cv)); trainY = Y(training(cv)); validationX = X(test(cv),:); validationAttribute = attribute(test(cv)); validationY = Y(test(cv));
Train a support vector machine (SVM) binary classifier on the training data. Standardize the predictors before fitting the model. Use the trained model to predict labels and compute scores for the validation data set.
mdl = fitcsvm(trainX,trainY,Standardize=true); [labels,scores] = predict(mdl,validationX);
For the validation data set, combine the sensitive attribute and response variable information into one grouping variable groupTest.
groupTest = validationAttribute.*validationY; names = string(categories(groupTest))
names = 4×1 string
"Female Smoker"
"Female Nonsmoker"
"Male Smoker"
"Male Nonsmoker"
Find the validation observations that are misclassified by the SVM model.
wrongIdx = (validationY ~= labels);
wrongX = validationX(wrongIdx,:);
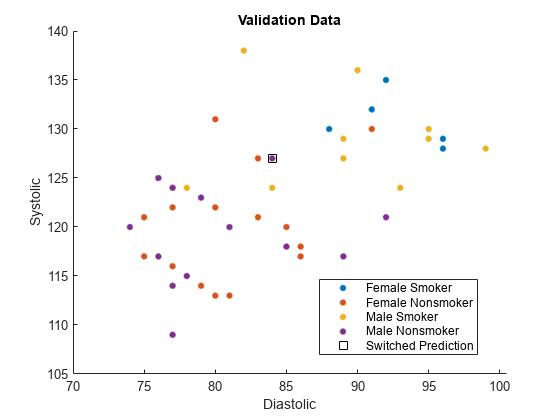
names(5) = "Misclassified";Plot the validation data. The color of each point indicates the sensitive attribute group and class label for that observation. Circled points indicate misclassified observations.
figure hold on gscatter(validationX(:,1),validationX(:,2), ... validationAttribute.*validationY) plot(wrongX(:,1),wrongX(:,2), ... "ko",MarkerSize=8) legend(names) xlabel("Diastolic") ylabel("Systolic") title("Validation Data") hold off
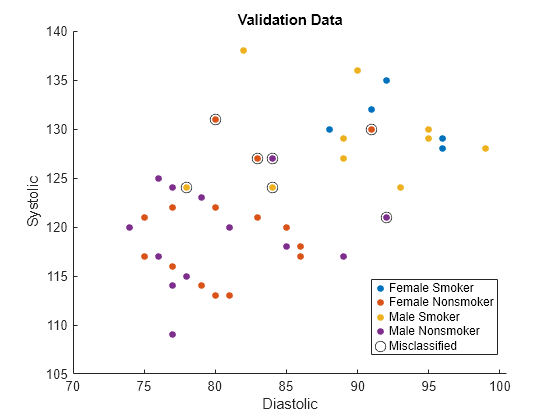
Compute fairness metrics with respect to the sensitive attribute by using the model predictions. In particular, find the statistical parity difference (SPD) for each group in validationAttribute.
metricsResults = fairnessMetrics(validationAttribute,validationY, ...
Predictions=labels);
metricsResults.ReferenceGroupans = 'Female'
metricsResults.PositiveClass
ans = categorical
Nonsmoker
report(metricsResults,BiasMetrics="StatisticalParityDifference")ans=2×4 table
ModelNames SensitiveAttributeNames Groups StatisticalParityDifference
__________ _______________________ ______ ___________________________
Model1 x1 Female 0
Model1 x1 Male -0.064412
figure
plot(metricsResults,"StatisticalParityDifference")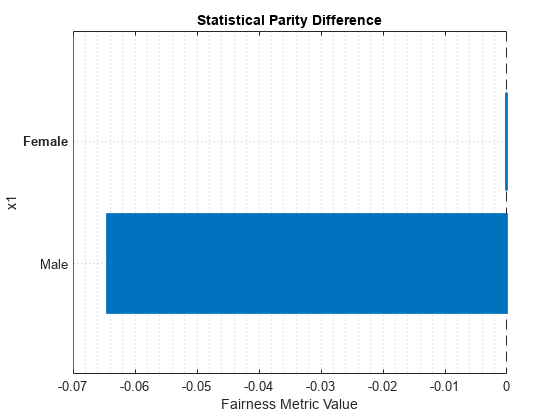
For the nonreference group (Male), the SPD value is the difference between the probability of a patient being in the positive class (Nonsmoker) when the sensitive attribute value is Male and the probability of a patient being in the positive class when the sensitive attribute value is Female (in the reference group). Ideally, SPD values are close to 0.
To try to improve the nonreference group SPD value, you can adjust the model predictions by using the fairnessThresholder function. The function searches for an optimal score threshold to maximize accuracy while satisfying fairness bounds. For observations in the critical region below the optimal threshold, the function changes the labels so that the fairness constraints hold for the reference and nonreference groups. By default, when you use the SPD bias metric, the function tries to find a score threshold such that the SPD value for the nonreference group is in the range [–0.05,0.05].
fairnessMdl = fairnessThresholder(mdl,validationX, ... validationAttribute,validationY, ... BiasMetric="StatisticalParityDifference")
fairnessMdl =
fairnessThresholder with properties:
Learner: [1×1 classreg.learning.classif.CompactClassificationSVM]
SensitiveAttribute: [50×1 categorical]
ReferenceGroups: Female
ResponseName: 'Y'
PositiveClass: Nonsmoker
ScoreThreshold: 0.5116
BiasMetric: 'StatisticalParityDifference'
BiasMetricValue: -0.0209
BiasMetricRange: [-0.0500 0.0500]
ValidationLoss: 0.1200
fairnessMdl is a fairnessThresholder model object.
Note that the updated nonreference group SPD value is closer to 0.
newNonReferenceSPD = fairnessMdl.BiasMetricValue
newNonReferenceSPD = -0.0209
Use the new score threshold to adjust the validation data predictions. The predict function of the fairnessMdl object adjusts the prediction of each observation whose maximum score is less than the score threshold. If the observation is in the nonreference group, the function predicts the observation into the positive class. If the observation is in the reference group, the function predicts the observation into the negative class. These adjustments do not always result in a change in the predicted label.
fairnessLabels = predict(fairnessMdl,validationX, ...
validationAttribute);Find the observations whose predictions are switched by fairnessMdl.
differentIdx = (labels ~= fairnessLabels);
differentX = validationX(differentIdx,:);
names(5) = "Switched Prediction";Plot the validation data. The color of each point indicates the sensitive attribute group and class label for that observation. Points in squares indicate observations whose labels are switched by the fairnessThresholder model.
figure hold on gscatter(validationX(:,1),validationX(:,2), ... validationAttribute.*validationY) plot(differentX(:,1),differentX(:,2), ... "ks",MarkerSize=8) legend(names) xlabel("Diastolic") ylabel("Systolic") title("Validation Data") hold off
Input Arguments
Fairness classification model, specified as a fairnessThresholder object. The ScoreThreshold property
of the object must be nonempty.
Data set, specified as a table. Each row of Tbl corresponds to
one observation, and each column corresponds to one variable. If you use a table when
creating the fairnessThresholder object, then you must use a table when
using the predict function. The table must include all required
predictor variables and the sensitive attribute. The table can include additional
variables, such as the response variable. Multicolumn variables and cell arrays other
than cell arrays of character vectors are not allowed.
Data Types: table
Predictor data, specified as a numeric matrix. Each row of X
corresponds to one observation, and each column corresponds to one predictor variable.
If you use a matrix when creating the fairnessThresholder object, then
you must use a matrix when using the predict function.
X and attribute must have the same number of
rows.
Data Types: single | double
Sensitive attribute, specified as a numeric column vector, logical column vector, character array, string array, cell array of character vectors, or categorical column vector.
Xandattributemust have the same number of rows.If
attributeis a character array, then each row of the array must correspond to a group in the sensitive attribute.
Data Types: single | double | logical | char | string | cell | categorical
Output Arguments
Class labels adjusted for fairness, returned as a numeric column vector, logical
column vector, character array, cell array of character vectors, or categorical column
vector. (The software treats a string scalar as
a character vector.) The function uses the data set predictions, adjusted
using the thresholder.ScoreThreshold value. For more information, see
Reject Option-Based Classification.
Version History
Introduced in R2023a
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)