t-SNE
What Is t-SNE?
t-SNE (tsne) is an algorithm for dimensionality reduction that is well-suited to visualizing high-dimensional data. The name stands for t-distributed Stochastic Neighbor Embedding. The idea is to embed high-dimensional points in low dimensions in a way that respects similarities between points. Nearby points in the high-dimensional space correspond to nearby embedded low-dimensional points, and distant points in high-dimensional space correspond to distant embedded low-dimensional points. (Generally, it is impossible to match distances exactly between high-dimensional and low-dimensional spaces.)
The tsne function creates a set of low-dimensional points from high-dimensional data. Typically, you visualize the low-dimensional points to see natural clusters in the original high-dimensional data.
The algorithm takes the following general steps to embed the data in low dimensions.
Calculate the pairwise distances between the high-dimensional points.
Create a standard deviation σi for each high-dimensional point i so that the perplexity of each point is at a predetermined level. For the definition of perplexity, see Compute Distances, Gaussian Variances, and Similarities.
Calculate the similarity matrix. This is the joint probability distribution of X, defined by Equation 1.
Create an initial set of low-dimensional points.
Iteratively update the low-dimensional points to minimize the Kullback-Leibler divergence between a Gaussian distribution in the high-dimensional space and a t distribution in the low-dimensional space. This optimization procedure is the most time-consuming part of the algorithm.
See van der Maaten and Hinton [1].
t-SNE Algorithm
The basic t-SNE algorithm performs the following steps.
Prepare Data
tsne first removes each row of the input data X that contains any NaN values. Then, if the Standardize name-value pair is true, tsne centers X by subtracting the mean of each column, and scales X by dividing its columns by their standard deviations.
The original authors van der Maaten and Hinton [1] recommend reducing the original data X to a lower-dimensional version using Principal Component Analysis (PCA). You can set the tsne
NumPCAComponents name-value pair to the number of dimensions you like, perhaps 50. To exercise more control over this step, preprocess the data using the pca function.
Compute Distances, Gaussian Variances, and Similarities
After the preprocessing, tsne calculates the distance d(xi,xj) between each pair of points xi and xj in X. You can choose various distance metrics using the Distance name-value pair. By default, tsne uses the standard Euclidean metric. tsne uses the square of the distance metric in its subsequent calculations.
Then for each row i of X, tsne calculates a standard deviation σi so that the perplexity of row i is equal to the Perplexity name-value pair. The perplexity is defined in terms of a model Gaussian distribution as follows. As van der Maaten and Hinton [1] describe, “The similarity of data point xj to data point xi is the conditional probability, , that xi would pick xj as its neighbor if neighbors were picked in proportion to their probability density under a Gaussian centered at xi. For nearby data points, is relatively high, whereas for widely separated data points, will be almost infinitesimal (for reasonable values of the variance of the Gaussian, σi).”
Define the conditional probability of j given i as
Then define the joint probability pij by symmetrizing the conditional probabilities:
| (1) |
where N is the number of rows of X.
The distributions still do not have their standard deviations σi defined in terms of the Perplexity name-value pair. Let Pi represents the conditional probability distribution over all other data points given data point xi. The perplexity of the distribution is
where H(Pi) is the Shannon entropy of Pi:
The perplexity measures the effective number of neighbors of point i. tsne performs a binary search over the σi to achieve a fixed perplexity for each point i.
Initialize the Embedding and Divergence
To embed the points in X into a low-dimensional space, tsne performs an optimization. tsne attempts to minimize the Kullback-Leibler divergence between the model Gaussian distribution of the points in X and a Student t distribution of points Y in the low-dimensional space.
The minimization procedure begins with an initial set of points Y. tsne create the points by default as random Gaussian-distributed points. You can also create these points yourself and include them in the 'InitialY' name-value pair for tsne. tsne then calculates the similarities between each pair of points in Y.
The probability model qij of the distribution of the distances between points yi and yj is
Using this definition and the model of distances in X given by Equation 1, the Kullback-Leibler divergence between the joint distribution P and Q is
For consequences of this definition, see Helpful Nonlinear Distortion.
Gradient Descent of Kullback-Leibler Divergence
To minimize the Kullback-Leibler divergence, the 'exact' algorithm uses a modified gradient descent procedure. The gradient with respect to the points in Y of the divergence is
where the normalization term
The modified gradient descent algorithm uses a few tuning parameters to attempt to reach a good local minimum.
'Exaggeration'— During the first 99 gradient descent steps,tsnemultiplies the probabilities pij from Equation 1 by the exaggeration value. This step tends to create more space between clusters in the output Y.'LearnRate'—tsneuses adaptive learning to improve the convergence of the gradient descent iterations. The descent algorithm has iterative steps that are a linear combination of the previous step in the descent and the current gradient.'LearnRate'is a multiplier of the current gradient for the linear combination. For details, see Jacobs [3].
Barnes-Hut Variation of t-SNE
To speed the t-SNE algorithm and to cut down on its memory usage, tsne offers an approximate optimization scheme. The Barnes-Hut algorithm groups nearby points together to lower the complexity and memory usage of the t-SNE optimization step. The Barnes-Hut algorithm is an approximate optimizer, not an exact optimizer. There is a nonnegative tuning parameter Theta that effects a tradeoff between speed and accuracy. Larger values of 'Theta' give faster but less accurate optimization results. The algorithm is relatively insensitive to 'Theta' values in the range (0.2,0.8).
The Barnes-Hut algorithm groups nearby points in the low-dimensional space, and performs an approximate gradient descent based on these groups. The idea, originally used in astrophysics, is that the gradient is similar for nearby points, so the computations can be simplified.
See van der Maaten [2].
Characteristics of t-SNE
Cannot Use Embedding to Classify New Data
Because t-SNE often separates data clusters well, it can seem that t-SNE can classify new data points. However, t-SNE cannot classify new points. The t-SNE embedding is a nonlinear map that is data-dependent. To embed a new point in the low-dimensional space, you cannot use the previous embedding as a map. Instead, run the entire algorithm again.
Performance Depends on Data Sizes and Algorithm
t-SNE can take a good deal of time to process data. If you have N data points in D dimensions that you want to map to Y dimensions, then
Exact t-SNE takes of order D*N2 operations.
Barnes-Hut t-SNE takes of order D*Nlog(N)*exp(dimension(Y)) operations.
So for large data sets, where N is greater than 1000 or so, and where the embedding dimension Y is 2 or 3, the Barnes-Hut algorithm can be faster than the exact algorithm.
Helpful Nonlinear Distortion
T-SNE maps high-dimensional distances to distorted low-dimensional analogues. Because of the fatter tail of the Student t distribution in the low-dimensional space, tsne often moves close points closer together, and moves far points farther apart than in the high-dimensional space, as illustrated in the following figure. The figure shows both Gaussian and Student t distributions at the points where the densities are at 0.25 and 0.025. The Gaussian density relates to high-dimensional distances, and the t density relates to low-dimensional distances. The t density corresponds to close points being closer, and far points being farther, compared to the Gaussian density.
t = linspace(0,5); y1 = normpdf(t,0,1); y2 = tpdf(t,1); plot(t,y1,'k',t,y2,'r') hold on x1 = fzero(@(x)normpdf(x,0,1)-0.25,[0,2]); x2 = fzero(@(x)tpdf(x,1)-0.25,[0,2]); z1 = fzero(@(x)normpdf(x,0,1)-0.025,[0,5]); z2 = fzero(@(x)tpdf(x,1)-0.025,[0,5]); plot([0,x1],[0.25,0.25],'k-.') plot([0,z2],[0.025,0.025],'k-.') plot([x1,x1],[0,0.25],'g-',[x2,x2],[0,0.25],'g-') plot([z1,z1],[0,0.025],'g-',[z2,z2],[0,0.025],'g-') text(1.1,.25,'Close points are closer in low-D') text(2.4,.05,'Far points are farther in low-D') legend('Gaussian(0,1)','Student t (df = 1)') xlabel('x') ylabel('Density') title('Density of Gaussian(0,1) and Student t (df = 1)') hold off
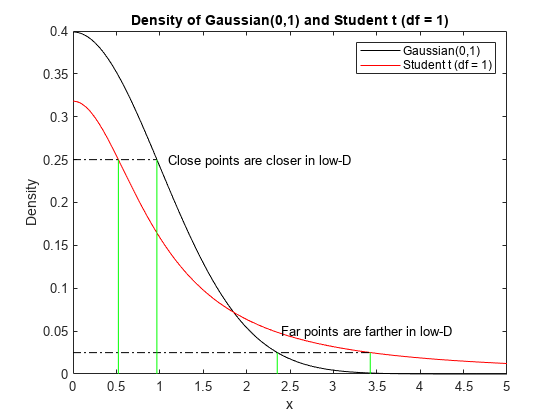
This distortion is helpful when it applies. It does not apply in cases such as when the Gaussian variance is high, which lowers the Gaussian peak and flattens the distribution. In such a case, tsne can move close points farther apart than in the original space. To achieve a helpful distortion,
Set the
'Verbose'name-value pair to2.Adjust the
'Perplexity'name-value pair so the reported range of variances is not too far from1, and the mean variance is near1.
If you can achieve this range of variances, then the diagram applies, and the tsne distortion is helpful.
For effective ways to tune tsne, see Wattenberg, Viégas and Johnson [4].
References
[1] van der Maaten, Laurens, and Geoffrey Hinton. "Visualizing Data using t-SNE." J. Machine Learning Research 9, 2008, pp. 2579–2605.
[2] van der Maaten, Laurens. Barnes-Hut-SNE. arXiv:1301.3342 [cs.LG], 2013.
[3] Jacobs, Robert A. "Increased rates of convergence through learning rate adaptation." Neural Networks 1.4, 1988, pp. 295–307.
[4] Wattenberg, Martin, Fernanda Viégas, and Ian Johnson. "How to Use t-SNE Effectively." Distill, 2016. Available at How to Use t-SNE Effectively.