wdenoise2
Wavelet image denoising
Syntax
Description
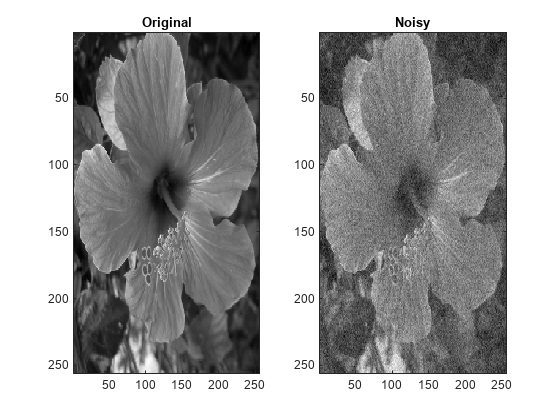
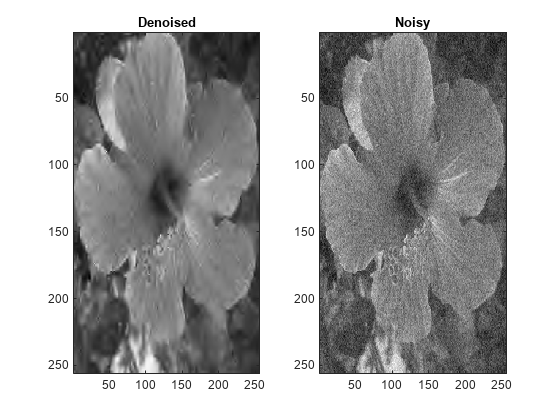
IMDEN = wdenoise2(IM)IM using an empirical Bayesian
method. The bior4.4 wavelet is used with a posterior median threshold
rule. Denoising is down to the minimum of floor(log2([M N])) and
wmaxlev([M N],'bior4.4'), where M and
N are the row and column sizes of the image.
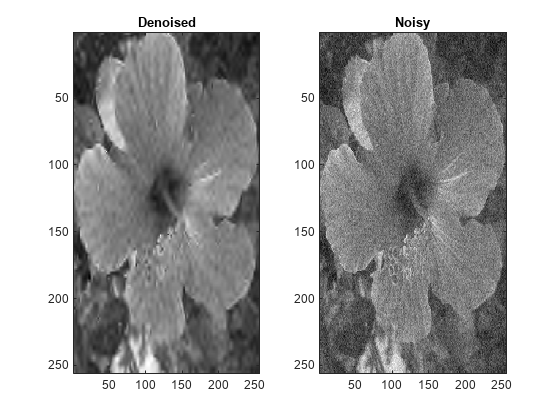
IMDEN is the denoised version of IM.
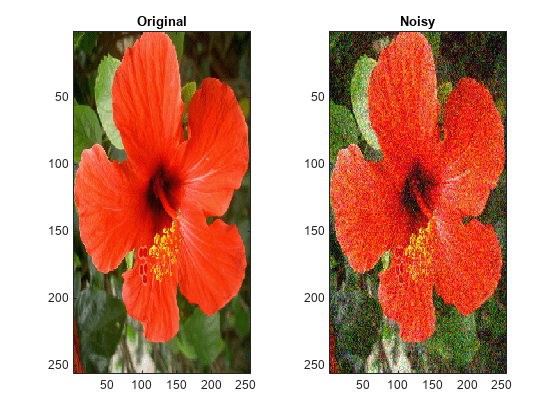
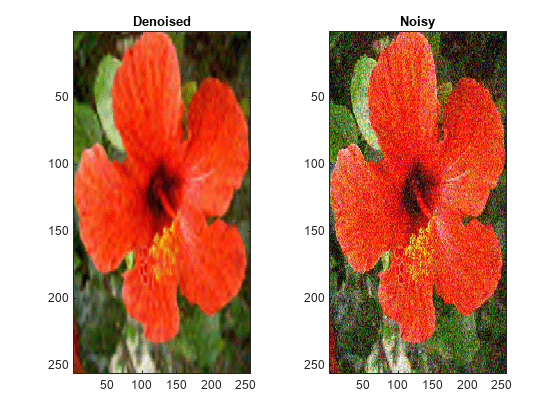
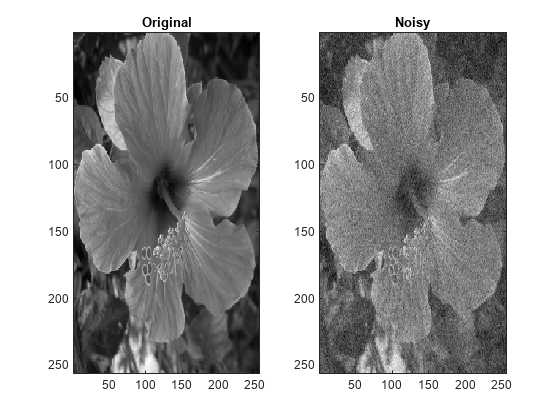
For RGB images, by default, wdenoise2 projects the image onto its
principal component analysis (PCA) color space before denoising. To denoise an RGB image
in the original color space, use the ColorSpace name-value
pair.
[
returns the scaling and denoised wavelet coefficients in IMDEN,DENOISEDCFS] = wdenoise2(___)DENOISEDCFS
using any of the preceding syntaxes.
[
returns the scaling and wavelet coefficients of the input image in
IMDEN,DENOISEDCFS,ORIGCFS] = wdenoise2(___)ORIGCFS using any of the preceding syntaxes.
[
returns the sizes of the approximation coefficients at the coarsest scale along with the
sizes of the wavelet coefficients at all scales. IMDEN,DENOISEDCFS,ORIGCFS,S] = wdenoise2(___)S is a matrix with
the same structure as the S output of wavedec2.
[
returns the shifts along the row and column dimensions for cycle spinning.
IMDEN,DENOISEDCFS,ORIGCFS,S,SHIFTS] = wdenoise2(___)SHIFTS is
2-by-(numshifts+1)2 matrix where each
column of SHIFTS contains the shifts along the row and column
dimension used in cycle spinning and numshifts is the value of
CycleSpinning.
[___] = wdenoise2(___,
returns the denoised image with additional options specified by one or more
Name,Value)Name,Value pair arguments, using any of the preceding
syntaxes.
wdenoise2(___) with no output arguments plots the
original image along with the denoised image in the current figure.
Examples
Input Arguments
Name-Value Arguments
Output Arguments
References
[1] Abramovich, F., Y. Benjamini, D. L. Donoho, and I. M. Johnstone. “Adapting to Unknown Sparsity by Controlling the False Discovery Rate.” Annals of Statistics, Vol. 34, Number 2, pp. 584–653, 2006.
[2] Antoniadis, A., and G. Oppenheim, eds. Wavelets and Statistics. Lecture Notes in Statistics. New York: Springer Verlag, 1995.
[3] Donoho, D. L. “Progress in Wavelet Analysis and WVD: A Ten Minute Tour.” Progress in Wavelet Analysis and Applications (Y. Meyer, and S. Roques, eds.). Gif-sur-Yvette: Editions Frontières, 1993.
[4] Donoho, D. L., I. M. Johnstone. “Ideal Spatial Adaptation by Wavelet Shrinkage.” Biometrika, Vol. 81, pp. 425–455, 1994.
[5] Donoho, D. L. “De-noising by Soft-Thresholding.” IEEE Transactions on Information Theory, Vol. 42, Number 3, pp. 613–627, 1995.
[6] Donoho, D. L., I. M. Johnstone, G. Kerkyacharian, and D. Picard. “Wavelet Shrinkage: Asymptopia?” Journal of the Royal Statistical Society, series B, Vol. 57, No. 2, pp. 301–369, 1995.
[7] Johnstone, I. M., and B. W. Silverman. “Needles and Straw in Haystacks: Empirical Bayes Estimates of Possibly Sparse Sequences.” Annals of Statistics, Vol. 32, Number 4, pp. 1594–1649, 2004.
Extended Capabilities
Version History
Introduced in R2019a