Proton Bragg Peak Fit
Feature
- Fit IDD curve with Bortfeld functions, IDD curve may contains multiple bragg peaks.
- Reconstruct 2D/3D proton dose from measurement of MLSIC[5], (multi-gauss model in XY plane, bortfeld in Z)
- Proton Radiography/CT with feature 1 and 2
- Provide IDD measurement data and simulation data, data acquired from Zebra and MLSIC which can be used in Proton Radiographics
- Bortfeld function implemented in C++, provide IDD, mean gradient and jacobian outputs.
- Input Integral Depth Dose(IDD) is suggested to rescale to [0,10] or nomalize to [0,1], but it's ok to fit raw data in any scale
- Compiled with Visual Studio + Intel OneAPI, faster than mex in MATLAB. Highly recommanded to compile the src with VS and Intel OneAPI
- Support Apple Silicon (Matlab R2022a, Matlab R2022a beta don't have toolboxs)
- Image Processing Toolbox, Optimization Toolbox and Signal Processing Toolbox required
bragg-peak fitting are ready to use.
Dose reconstruction Working in progress
How to use:
examples: ./utils/demo.m
Windows
- Pre-compiled functions are provided.
-
BortfeldFunction.mexw64Windows with avx2(supported by most of modern x86_64 CPU).
-
- If Pre-compiled functions not work, run
./src/compile_PBPF.mto compile it
MacOS
- Pre-compiled functions are provided.
-
BortfeldFunction.mexmaca64for Apple Silicon Mac (do not Support AVX) -
BortfeldFunction.mexmacai64for Intel Mac (AVX not used)
-
- If Pre-compiled functions not work, run
./src/compile_PBPF.mto compile it
Linux
- Pre-compiled functions are Not provided.
-
BortfeldFunction_avx512f.mexa64Linux with avx512f (Only some x86_64 CPU support this) If you use this, change BortfeldFunction in bf_mex to BortfeldFunction_avx512f -
BortfeldFunction.mexa64Linux with avx2(supported by most of modern x86_64 CPU)
-
- If Pre-compiled functions not work, run
./src/compile_PBPF.mto compile it
Compilation
- All precompiled mex function can work with both
singleanddouble, butsinglenumbers can not be used for finite difference optimization(double precision required). Use the gradient provided by mex function. - Open the root folder run
cd('./src')in matlab - run
compile_PBPFthree compile flags are provided- use_openmp = 1; Use OpenMP
- use_avx = 1; Use AVX2
- is_avx512 = 1; Use AVX512 (not every CPU support this)
Here are some code snippets
- predict IDD with depth and bf parameters
z = (1:64)*0.3; % depth unit(cm), (n,1) size is preferred
% bortfeld function parameters, [range1,sigma1,epsilon1,phi1, range2,sigma2,epsilon2,phi2] two bragg-peaks used here, range unit is cm
bf_para = [15,0.3,1e-3,0.4, 12,0.4,1e-3,0.4];
% get idd curve, size of output will be (n,1), no matter z is (1,n) or (n,1)
output = bf_mex(z,bf_para,'idd');
plot(z,output);
grid on;
grid minor;
xlabel('Depth(cm)');
ylabel('Dose(a.u.)');% a.u. means arbitary unit
% for short
% output = bf_mex((1:64)*0.3,[15,0.3,1e-3,0.4, 12,0.4,1e-3,0.4],'idd')- Fit measured IDD
% method 1
[x,idd_o] = precise_fit(z,idd_i,num_bp,strict);
% method 2 used for fast fit, don't calculate IDD in this function
x = fast_fit(z,idd_i,num_bp);
idd_o = bf_mex(z,x,'idd');- Get gradient (Jacobian matrix) of Bortfeld function
z = (1:64)*0.3; % depth unit(cm), (n,1) size is preferred
% bortfeld function parameters, (m,1), range unit is cm
bf_para = [15,0.3,1e-3,0.4, 12,0.4,1e-3,0.4];
% output size will be (n,m)
output = bf_mex((1:64)'*0.3,[15,0.3,1e-3,0.4, 12,0.4,1e-3,0.4],'jacobian')- Other functions
- stored in
./utils/ - If matlab crashed on Rosseta Matlab, run this function
ifMacCrashed()
- stored in
- Benchmark of mex functions
./utils/test_mex_func.m, further optimization required.
% BortfeldFunction time: single(0.249554s), double(0.248533s)
% BortfeldFunction Grad time: single(2.758517s), double(2.772066s)
% Gauss2D Grad GPU time: single(0.942952s), double(1.575574s)
% Gauss2D Grad CPU time: single(0.131198s), double(0.201214s)
% Gauss2D GPU time: single(0.196284s), double(0.450232s)
% Gauss2D CPU time: single(0.842755s), double(0.665228s)
% ProtonDose3D GPU time: single(0.493263s), double(0.537123s)
% ProtonDose3D CPU time: single(0.751850s), double(0.496489s)s5cuGAUFbX1a
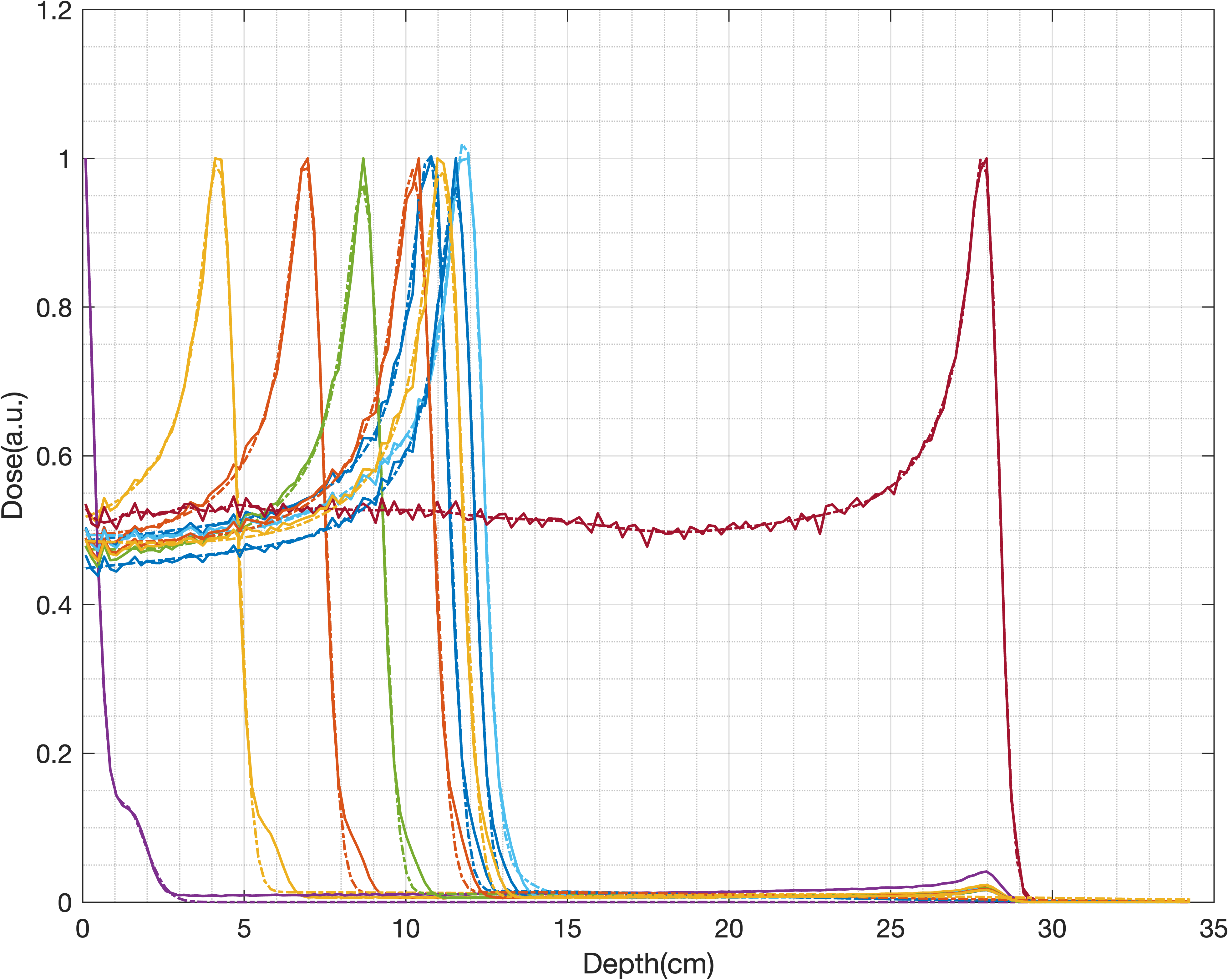
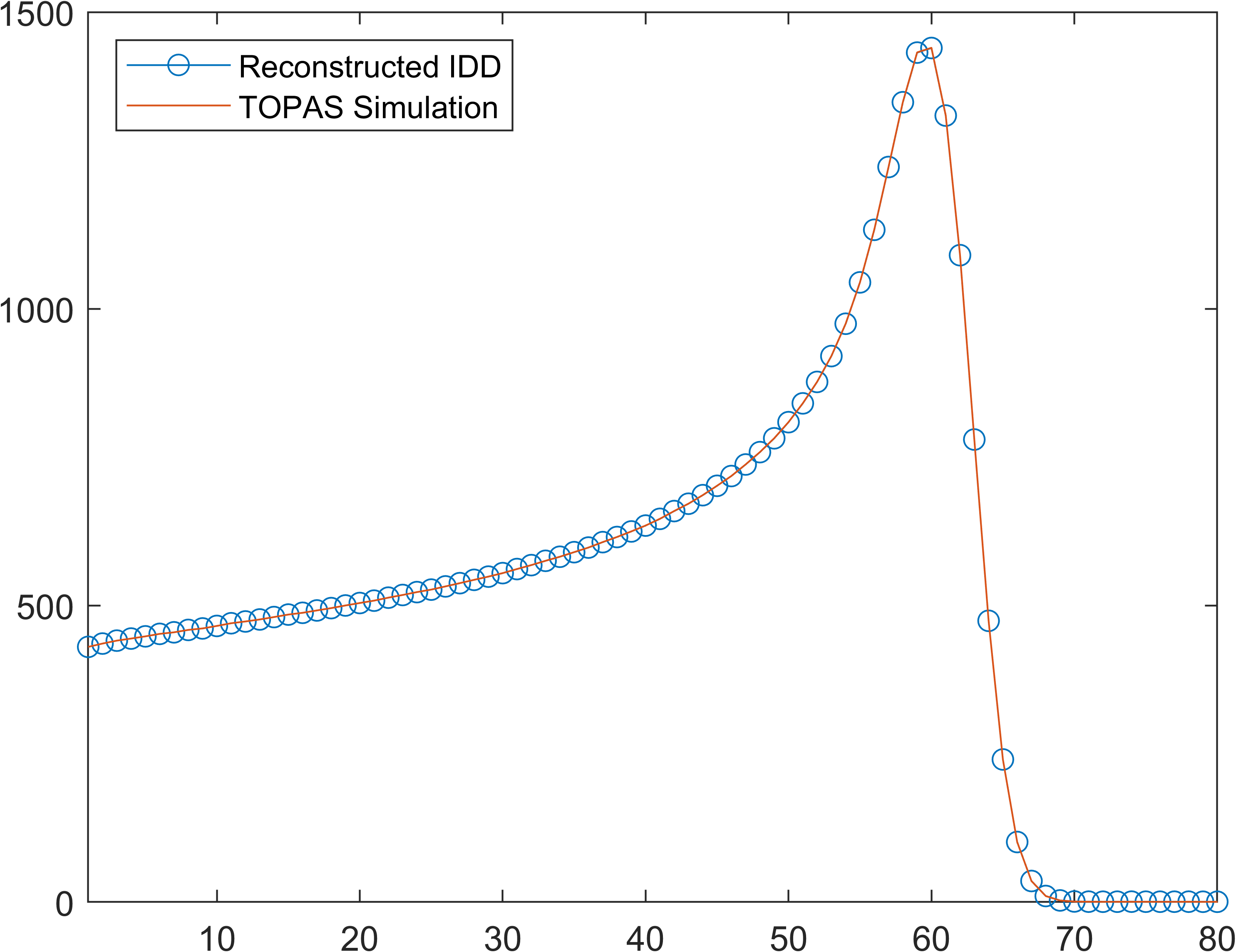
1D IDD data
Applications can be found in ./utils/demo.m and data stored in ./data/. Be careful, run the demo section by section, some parts are time comsuming.
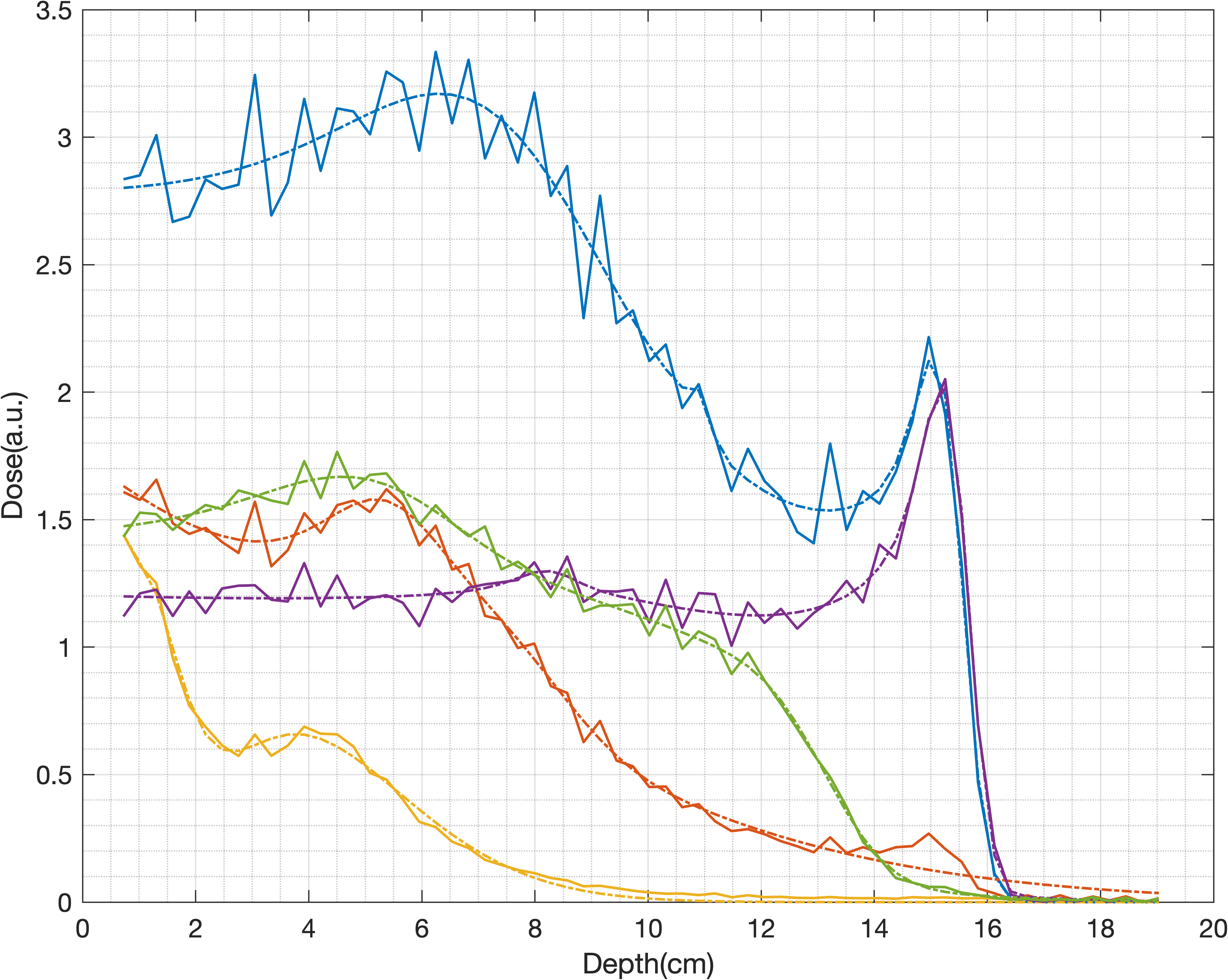
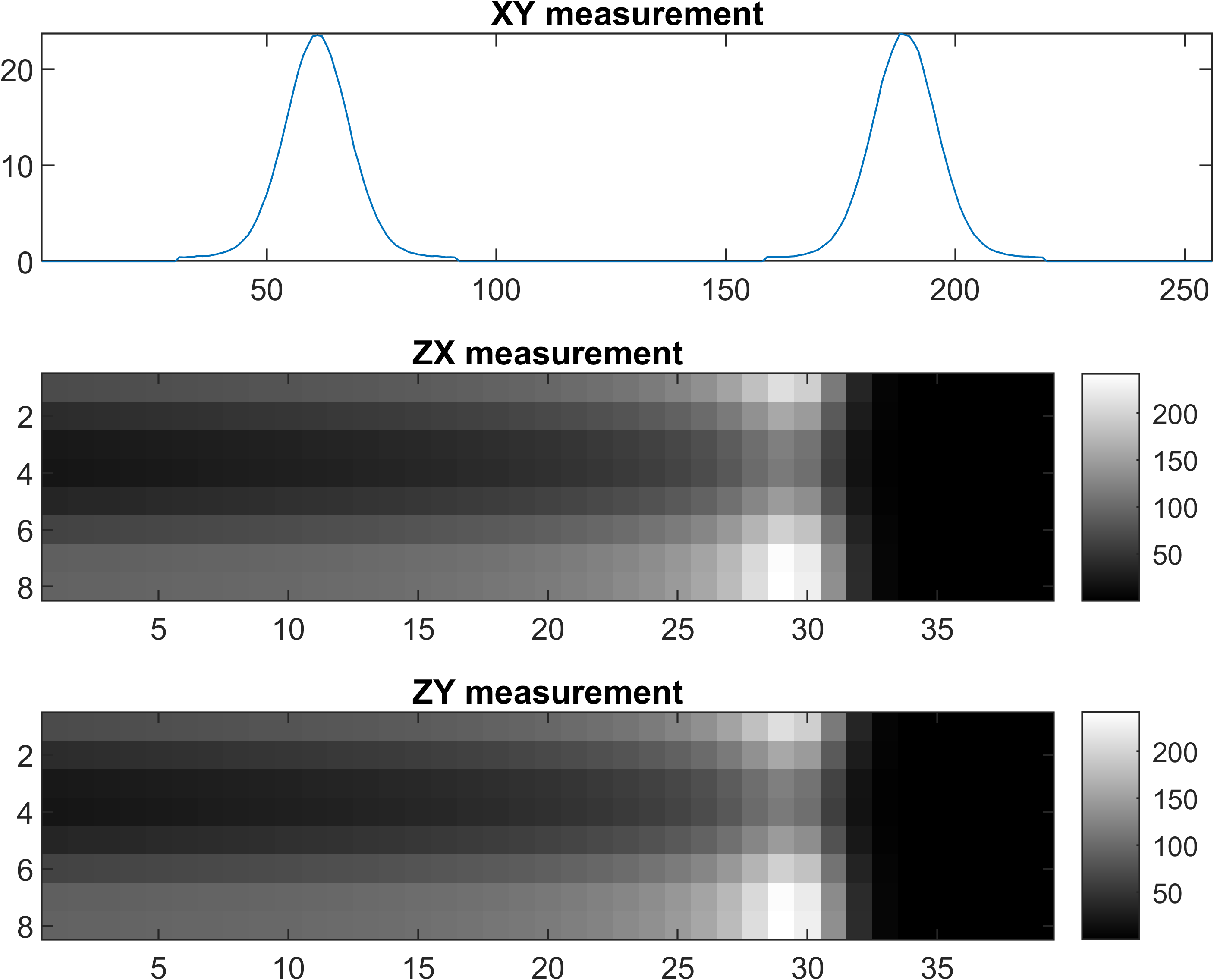
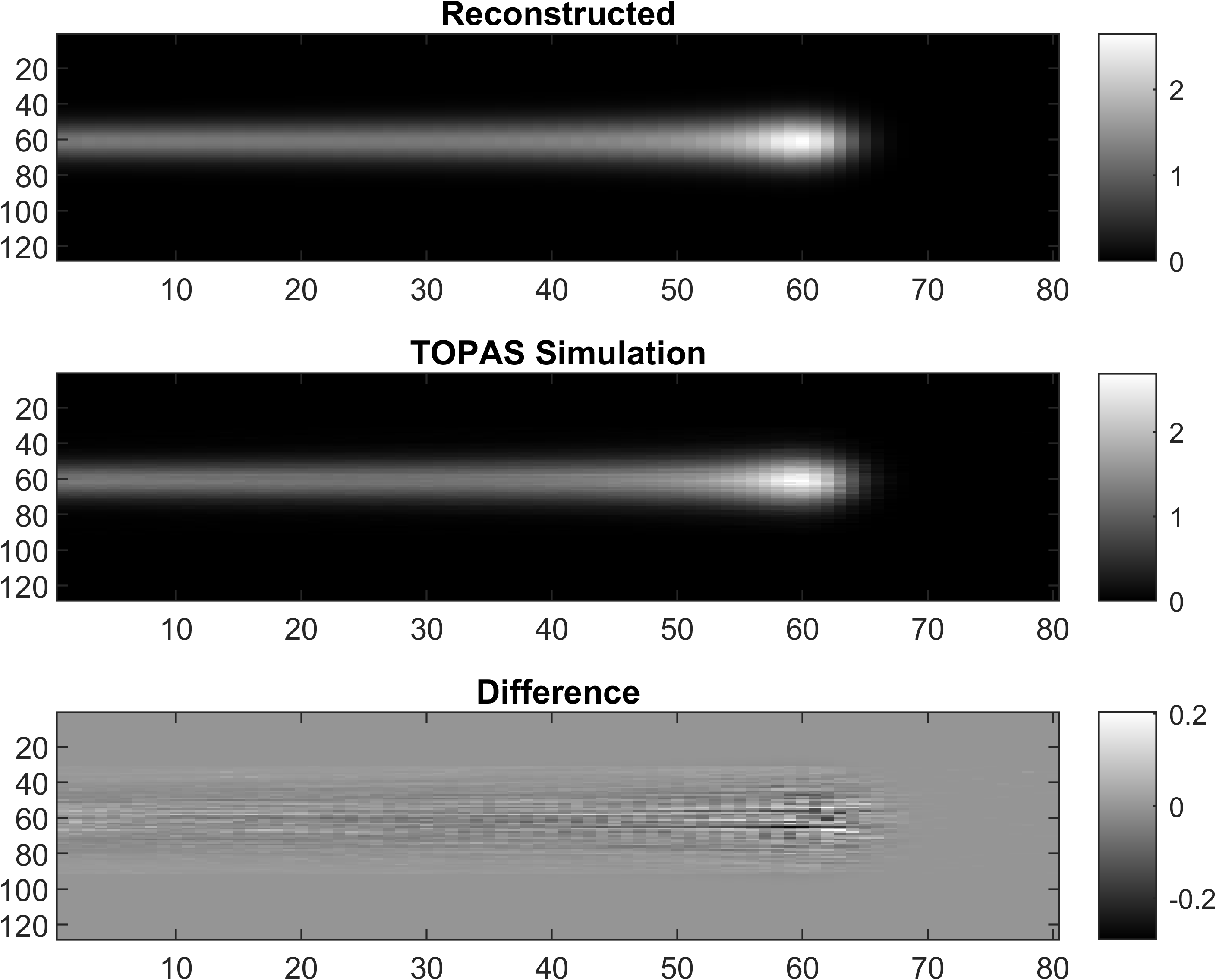
2D dose reconstruction
Strip ionization chamber array[4,5] were used for QA purpose, two layers of horizontal and vertical strip detectors were used to locate the proton beam position.Details
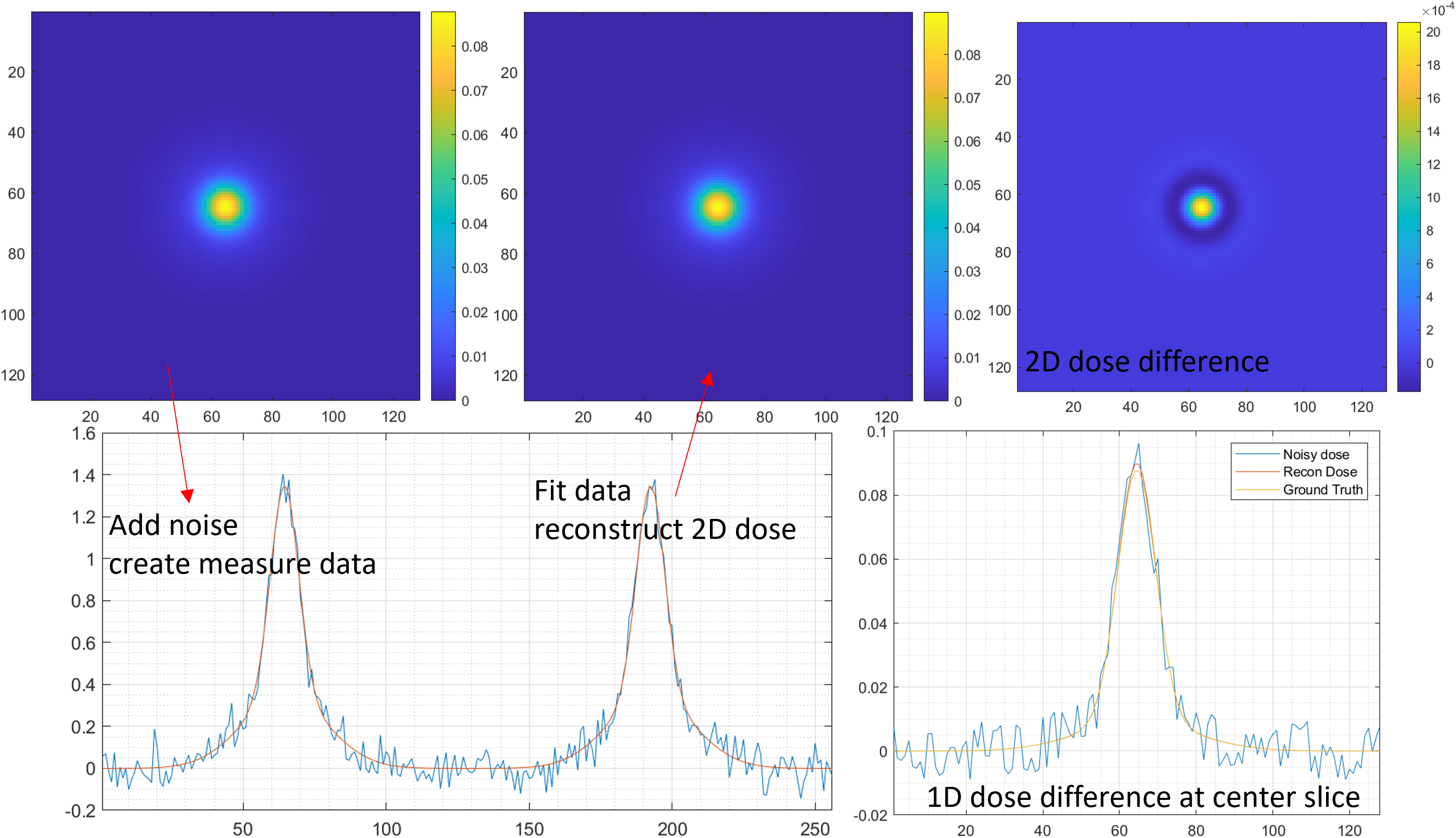
| White Noise level(divide by max dose) | Max Absolute Error( <math-renderer class="js-inline-math" style="display: inline" data-static-url="https://github.githubassets.com/static" data-run-id="48b536abf7cf51b9a6cde2f0de740e48">$\frac{\max|error|}{max[dose]}$</math-renderer>) |
|---|---|
| 30% | 0.0441 |
| 20% | 0.0337 |
| 10% | 0.0192 |
3D dose reconstruction
Only MLSIC can measure a 3D dose in real time[5], it contains 768 ADC channels (XY: 128x2, ZX: 8x32, ZY: 8x32) Details. The measurement function of MLSIC can be defined as a function, <math-renderer class="js-inline-math" style="display: inline" data-static-url="https://github.githubassets.com/static" data-run-id="48b536abf7cf51b9a6cde2f0de740e48">$f: \R^{128\times128\times66} \rightarrow \R^{128\times2 + 8\times32 + 8\times32}$</math-renderer>
Symmetric dose
Asymetric dose
to be updated
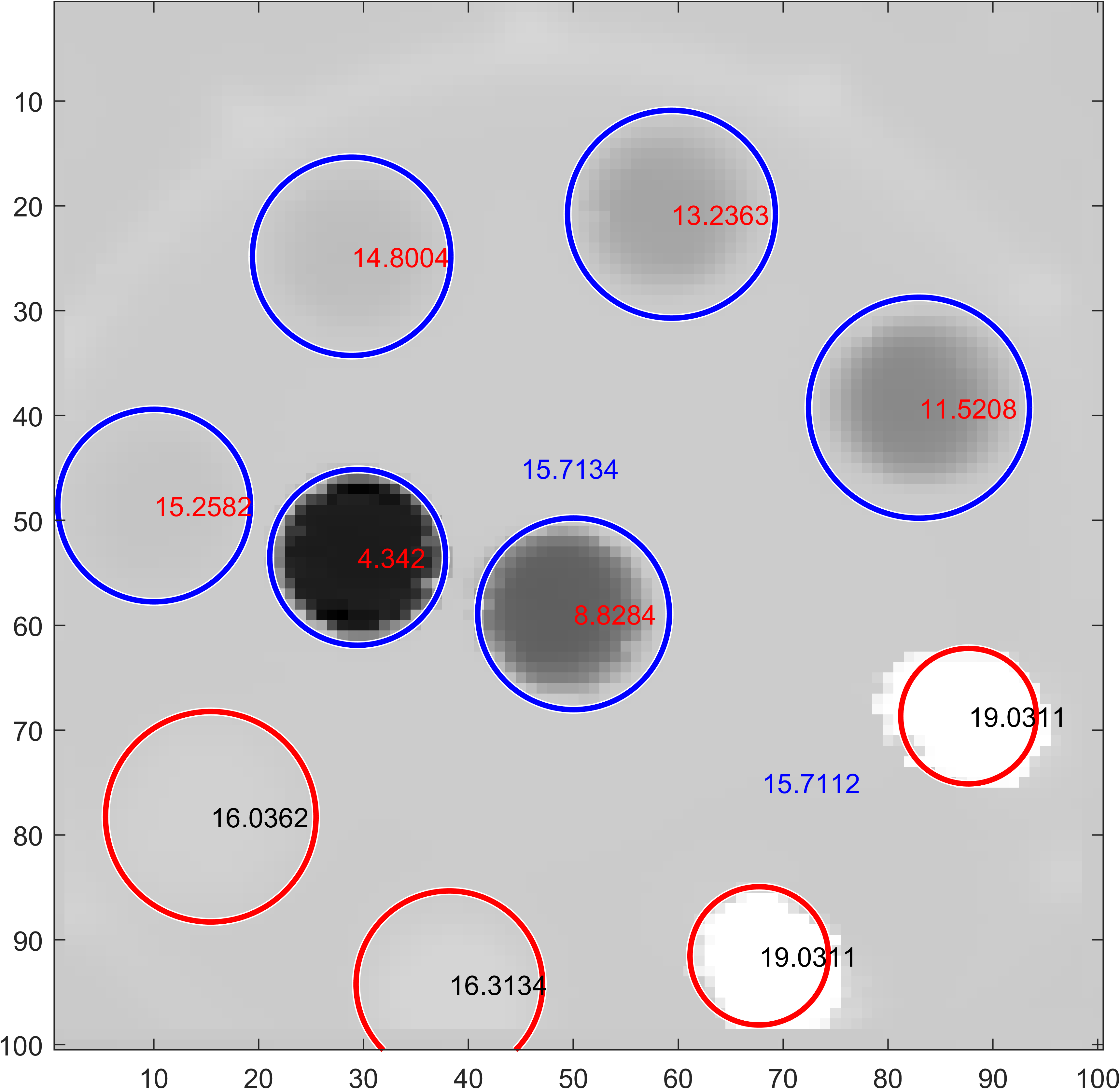
Proton Radiography
| Head Phantom | Simulation | MLSIC data |
|---|---|---|
 |
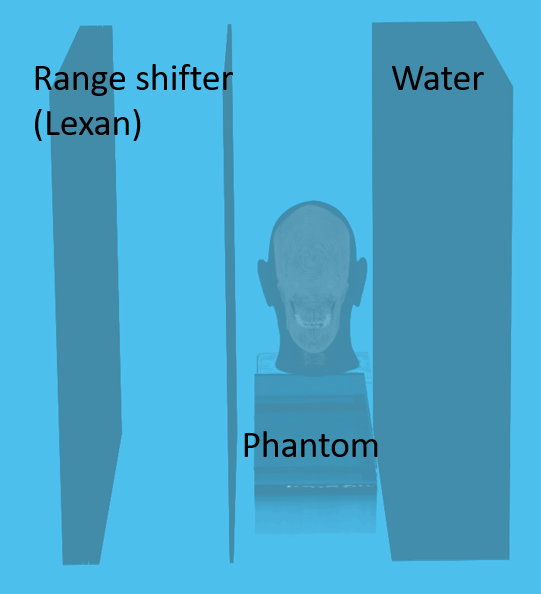 |
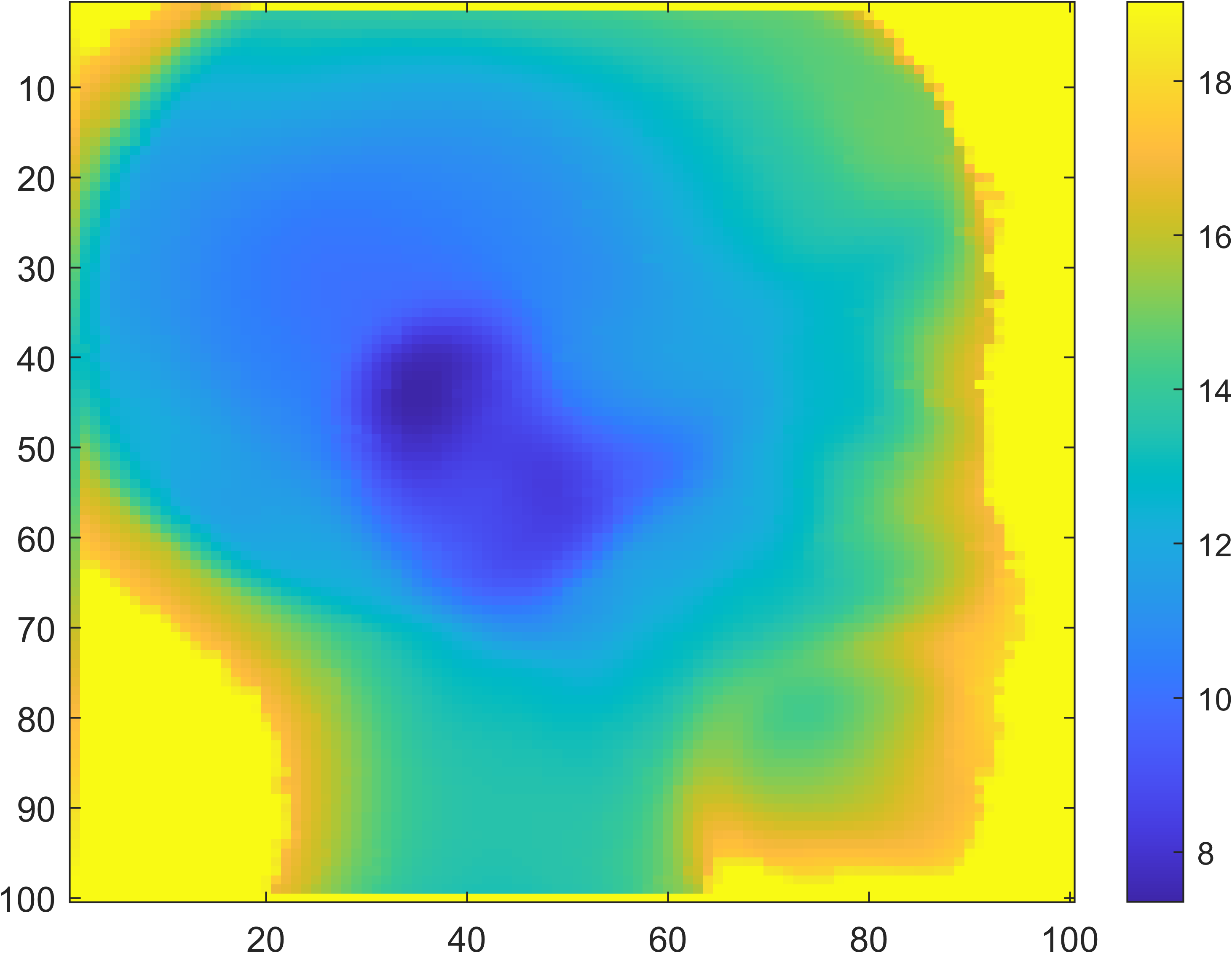 |
Proton CT
to be updated
Details of math and MLSIC structures can be found in here
Reference :
- An analytical approximation of the Bragg curve for therapeutic proton beams
- E. Cojocaru. Parabolic Cylinder Functions
- On the parametrization of lateral dose profiles in proton radiation therapy
- A 2D strip ionization chamber array with high spatiotemporal resolution for proton pencil beam scanning FLASH radiotherapy
- A multi-layer strip ionization chamber (MLSIC) device for proton pencil beam scan quality assurance
- Evaluation of the influence of double and triple Gaussian proton kernel models on accuracy of dose calculations for spot scanning technique
Remark:
M1 mac openmp issue: https://www.mathworks.com/matlabcentral/answers/1761950-m1-mac-compile-mex-file-with-openmp?s_tid=srchtitle
Matlab R2022a thread safty is not very stable, if the mex function crashed matlab, run the magic function 'test3_mex.mexmaci64' by calling test3_mex(int32(16));. Its output looks like
i = 1
i = 2
i = 7
i = 8
i = 5
i = 6
i = 3
i = 4
i = 9
i = 10
i = 13
i = 11
i = 14
i = 15
i = 12
i = 16
Matlab R2022a beta works well with openmp but it do not support official toolbox.
引用格式
shuang zhou (2025). Fit Proton Bragg Peaks (https://github.com/civerjia/Proton-Bragg-Peak-Fit/releases/tag/v1.3.0), GitHub. 检索时间: .
MATLAB 版本兼容性
平台兼容性
Windows macOS Linux标签
Community Treasure Hunt
Find the treasures in MATLAB Central and discover how the community can help you!
Start Hunting!src
utils
| 版本 | 已发布 | 发行说明 | |
|---|---|---|---|
| 1.3.0 | See release notes for this release on GitHub: https://github.com/civerjia/Proton-Bragg-Peak-Fit/releases/tag/v1.3.0 |
||
| 1.2.1 | See release notes for this release on GitHub: https://github.com/civerjia/Proton-Bragg-Peak-Fit/releases/tag/v1.2.1 |
||
| 1.2.0 | See release notes for this release on GitHub: https://github.com/civerjia/Proton-Bragg-Peak-Fit/releases/tag/v1.2.0 |
||
| 1.1.0 | See release notes for this release on GitHub: https://github.com/civerjia/Proton-Bragg-Peak-Fit/releases/tag/v1.1.0 |
||
| 1.0.1 | add cpp files |
||
| 1.0.0 |