SRAFasterqDumpOptions
Description
An SRAFasterqDumpOptions object contains options for the
srafasterqdump
function, which you use to download the files from SRA (Sequence Read Archive) [1].
Creation
Syntax
Description
sraOpt = SRAFasterqDumpOptionsSRAFasterqDumpOptions object with default property values.
SRAFasterqDumpOptions requires the SRA Toolkit for Bioinformatics Toolbox™. If this support package is not installed, then the function provides a download
link. For details, see Bioinformatics Toolbox Software Support Packages.
sraOpt = SRAFasterqDumpOptions(Name=Value)SRAFasterqDumpOptions object, specify
IncludeTechnical=true to set the value of the
IncludeTechnical property to true, so that
technical reads are included in the downloaded files.
sraOpt = SRAFasterqDumpOptions(S)S.
Input Arguments
srafasterqdump options, specified as a character vector or
string scalar. S must be in the original
fasterq-dump option syntax (prefixed by one or two
dashes).
Example: "--fasta-ref-tbl --internal-ref"
Properties
Flag to append new data to the output file instead of overwriting it, specified as a numeric
or logical 1 ( true) or 0 (false). By default, the
output file is overwritten with new data.
Data Types: double | logical
Flag to concatenate sequence information pertaining to each spot, specified as a
numeric or logical 1 (true) or 0 (false). By
default, the software does not concatenate the information pertaining to each spot. That
is, the software writes four lines of FASTQ or two lines of FASTA into one output file
for each spot. For details, see FASTQ/FASTA concatenated.
Data Types: double | logical
Additional commands, specified as a character vector or string scalar.
The commands must be in the native syntax (prefixed by one or two dashes). Use this option to apply undocumented flags and flags without corresponding MATLAB® properties.
Example: ExtraCommand="--fasta-ref-tbl --internal-ref"
Data Types: char | string
Flag to save the output in the FASTA format, specified as a numeric or logical
1 (true) or 0 (false). The default
output format is the FASTQ format.
Data Types: double | logical
Flag to split sequence information pertaining to each spot without
preserving the spot order, specified as a numeric or logical 1 (true)
or 0 (false).
If the value is true, the software splits the sequence information
in each spot is into reads. For each read, two lines of FASTA are written into the
single output file. Setting FastaOutputUnsorted=true is the same as
setting SplitType=SplitSpot, with the following exceptions:
With
FastaOutputUnsorted=true, the original order of the spots and reads is not preserved, andFastaOutputUnsortedname-value argument is exclusively for the FASTA output.This setting is faster than the
SplitSpotoption and does not use temporary files.
Data Types: double | logical
String of bases used to filter the output, specified as a string scalar. The output is filtered by comparing it to the specified string of bases and keeping reads that include the specified string of bases.
Data Types: string
Flag to include all object properties with
corresponding default values when converting properties to the original option syntax,
specified as a numeric or logical 1 (true) or 0
(false). You can convert properties to the original syntax
prefixed by one or two dashes (such as '-e 8 --split-file') by using
the getCommand function.
When IncludeAll=false and you call
getCommand(optionsObject), the software converts only the
specified properties. If the value is true,
getCommand converts all available properties, using default
values for unspecified properties, to the original syntax.
Note
If you set IncludeAll to true, the software
converts all available properties, using default values for unspecified properties. The
only exception is when the default value of a property is NaN,
Inf, [], '', or
"". In this case, the software does not translate the
corresponding property.
Data Types: logical | double
Flag to include technical reads in the downloaded files,
specified as a numeric or logical 1
(true) or 0
(false).
Data Types: double | logical
Minimum length required for a read to be included in the output, specified as a nonnegative integer. By default, no read is filtered out.
Data Types: double
Number of parallel threads to use, specified as a positive integer. The software runs threads on separate processors or cores. Increasing the number of threads generally improves the runtime significantly, but also increases the memory footprint.
Data Types: double
Folder where the output files are saved, specified as a character vector or string scalar. By default, the software saves the files in the current directory.
Data Types: char | string
Base name of the output files, specified as a character vector or string scalar. The default base name is the accession run number.
Data Types: char | string
Method used to split sequence information pertaining to each spot, specified as one of the following:
"SplitThree"— The software splits spots into reads. For each read, the software writes four lines of FASTQ or two lines of FASTA. For spots with two reads, the software produces*_1.fastqand*_2.fastqfiles. The software places unmated reads in*.fastq. If the accession does not have any spot with one single read, the software does not create a*.fastqfile. For details, see FASTQ/FASTA split 3."SplitSpot"— The software splits spots into reads. For each read, the software writes four lines of FASTQ or two lines of FASTA. All the reads are saved to a single output file. For details, see FASTQ/FASTA split spot."SplitFiles"— The software splits spots into reads. For each read, the software writes four lines of FASTQ or two lines of FASTA. The software assigns each read a number n, where 1 ≤ n ≤ 5, and then saves each nth read to the nth file (*_n.fastq). For details, see FASTQ/FASTA split file.
By default, the reads refer to biological reads only. However, if you set
IncludeTechnical to true, then the software
also includes the technical reads in the output files.
Data Types: char | string
This property is read-only.
Supported version of the original fasterq-dump software, returned as a
string scalar.
Data Types: string
Object Functions
getCommand | Translate object properties to original options syntax |
getOptionsTable | Return table with all properties and equivalent options in original syntax |
Examples
Download some paired-end sequencing data in a FASTQ format using an accession run number SRR11846824 that has two reads per spot and has no unaligned reads. Downloading the data may take a few minutes.
tbl = srafasterqdump("SRR11846824")tbl=1×2 table
Reads_1 Reads_2
_____________________ _____________________
SRR11846824 "SRR11846824_1.fastq" "SRR11846824_2.fastq"
By default, the function uses the SplitType="SplitThree" option and downloads only biological reads. Specifically, the function splits spots into reads. For spots having two reads, the function produces *_1.fastq and *_2.fastq, represented by the Reads_1 and Reads_2 columns. If there are any unaligned reads, the function saves unaligned reads in a *.fastq file, which would be represented by the Reads column. Because there are no unaligned reads within this accession, the function did not produce a *.fastq file, and the output table has no Reads column. For details, see SplitType.
You can also specify other download options using SRAFasterqDumpOptions. For instance, use FastaOutput=true to get the FASTA-formatted file.
sraopt = SRAFasterqDumpOptions;
sraopt.FastaOutput = true;
tbl2 = srafasterqdump("SRR11846824",sraopt);Alternatively, you can specify the options as name-value arguments instead of using the options object.
tbl2 = srafasterqdump("SRR11846824",FastaOutput=true);
You can also download the data in a SAM format using srasamdump.
samFile = srasamdump("SRR11846824")samFile = "SRR11846824.sam"
Specify the download options using an SRASAMDumpOptions object. For instance, specify the output file name and compress the output file using bzip2.
samdumpopt = SRASAMDumpOptions;
samdumpopt.OutputFileName = "SRR11846824.sam.bz2";
samdumpopt.BZip2 = truesamdumpopt =
SRASAMDumpOptions with properties:
Default properties:
ExtraCommand: ""
FastaOutput: 0
FastqOutput: 0
GZip: 0
HideIdentical: 0
IncludeAll: 0
MinMapQuality: 0
OutputPrimary: 0
OutputUnaligned: 0
Version: "3.0.6"
Modified properties:
OutputFileName: "SRR11846824.sam.bz2"
BZip2: 1
bzFile = srasamdump("SRR11846824",samdumpopt)bzFile = "SRR11846824.sam.bz2"
After downloading the SAM file, you can use it for downstream analyses. For instance, you can use bowtie2 to map the reads to the reference sequence.
First, download the C. elegans reference sequence.
celegans_refseq = fastaread("https://s3.amazonaws.com/igv.broadinstitute.org/genomes/seq/ce11/ce11.fa");Save Chromosome 3 reference data in a FASTA file.
celegans_chr3 = celegans_refseq(3).Sequence; warnState = warning; warning('off','Bioinfo:fastawrite:AppendToFile'); fastawrite("celegans_chr3.fa",celegans_chr3); warning(warnState);
Build a set of index files using bowtie2build. The status value of 0 means that the build was successful.
status = bowtie2build("celegans_chr3.fa","celegans_chr3_index");
Align read data to the reference. This may take a few minutes.
bowtie2("celegans_chr3_index","SRR11846824_1.fastq","SRR11846824_2.fastq","SRR11846824_mapped.sam");
Create a quality control plot for the SAM file. Note that, for this particular experiment, most of the reads happen to have the same quality score of 30.
seqqcplot("SRR11846824_mapped.sam");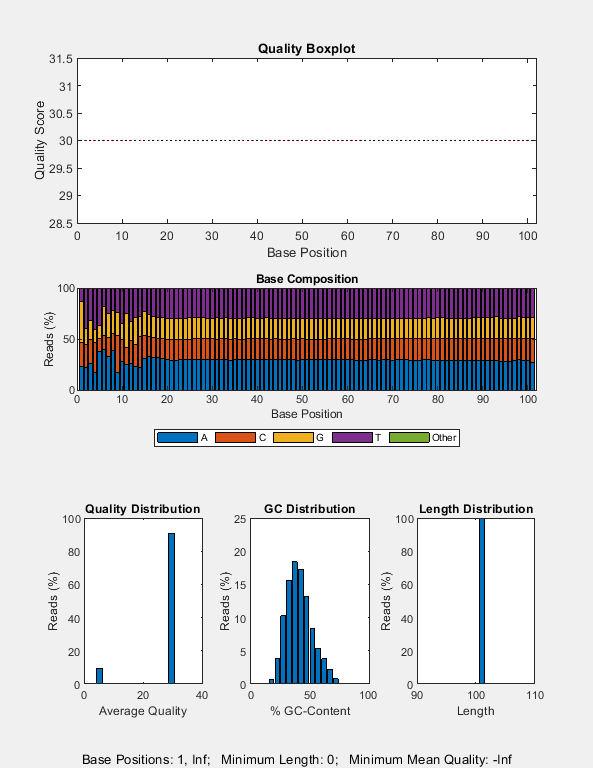
Convert the SAM file to a BAM file. Suppress two informational warnings that are issued while creating a BioMap object.
w = warning; warning("off","bioinfo:BioMap:BioMap:UnsortedReadsInSAMFile"); warning("off","bioinfo:saminfo:InvalidTagField"); bmObj = BioMap("SRR11846824_mapped.sam"); write(bmObj,"SRR11846824_mapped.bam",Format="BAM"); warning(w);
Visualize the alignment data in the Genomics Viewer app. The corresponding cytoband file is provided with the toolbox.
gv = genomicsViewer(ReferenceFile="celegans_chr3.fa",CytoBand="celegans_cytoBandIdeo.txt.gz"); addTracks(gv,"SRR11846824_mapped.bam");
Use the zoom slider to zoom in and see the features. Or you can enter the following in the search text box: Generated:3,711,861-3,711,940.
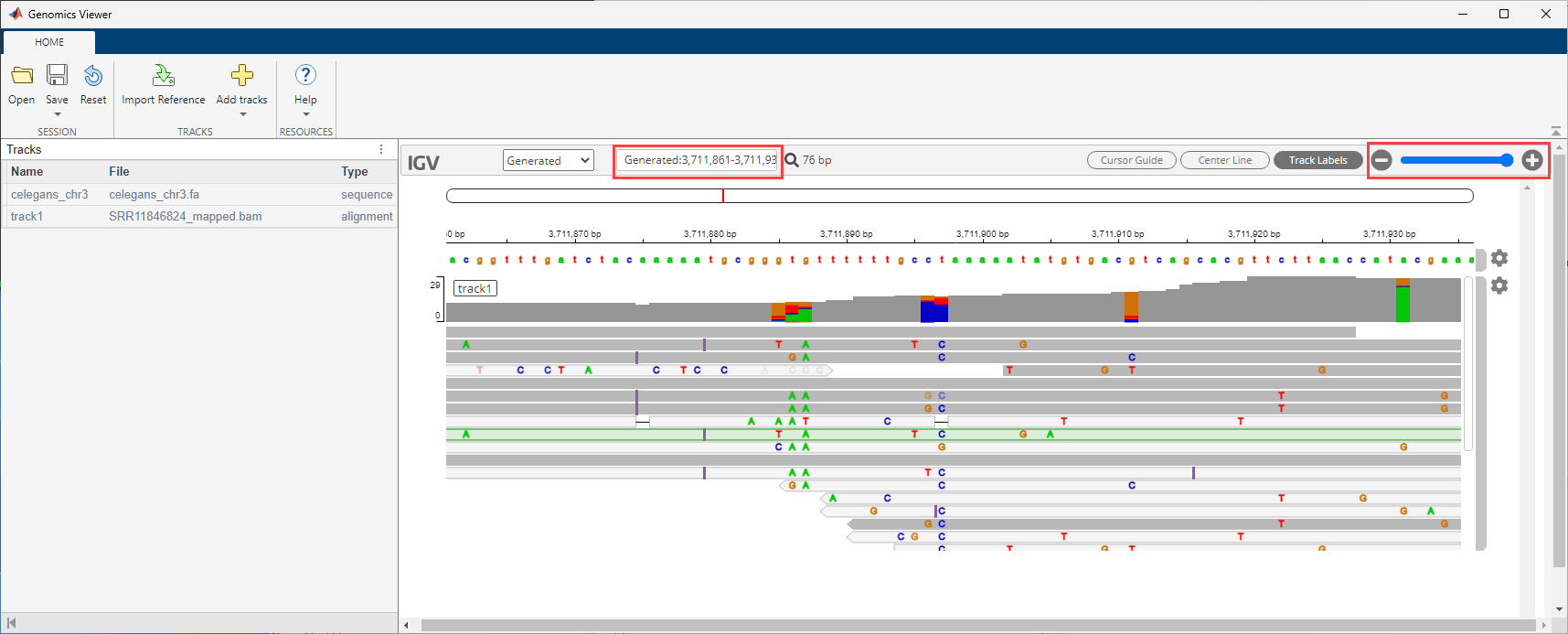
You may delete the downloaded files, such as the reference sequence file.
delete celegans_chr3.faClose the app.
close(gv);
References
[1] SRA Toolkit Development Team https://github.com/ncbi/sra-tools/wiki/01.-Downloading-SRA-Toolkit
Version History
Introduced in R2024a
See Also
MATLAB Command
You clicked a link that corresponds to this MATLAB command:
Run the command by entering it in the MATLAB Command Window. Web browsers do not support MATLAB commands.
选择网站
选择网站以获取翻译的可用内容,以及查看当地活动和优惠。根据您的位置,我们建议您选择:。
您也可以从以下列表中选择网站:
如何获得最佳网站性能
选择中国网站(中文或英文)以获得最佳网站性能。其他 MathWorks 国家/地区网站并未针对您所在位置的访问进行优化。
美洲
- América Latina (Español)
- Canada (English)
- United States (English)
欧洲
- Belgium (English)
- Denmark (English)
- Deutschland (Deutsch)
- España (Español)
- Finland (English)
- France (Français)
- Ireland (English)
- Italia (Italiano)
- Luxembourg (English)
- Netherlands (English)
- Norway (English)
- Österreich (Deutsch)
- Portugal (English)
- Sweden (English)
- Switzerland
- United Kingdom (English)